Run an experiment - syue99/Lab_control GitHub Wiki
In this page we talk about the logic and steps when we run a real experiment using the Labrad. Please check out playground for a more fundamental introduction on how to different clients and servers are used in labrad to do a simple toy experiment.
Backgrounds
Clients and Servers: A recap in the experiment prospective
Different than the toy experiment with a minimalistic fashion of the servers, a real experiment often wants a variety of servers and clients. Of course we eventually need to use all servers and clients in various experiments, but a rough classification of the "Core" servers that basically needs in every experiment include the followings:
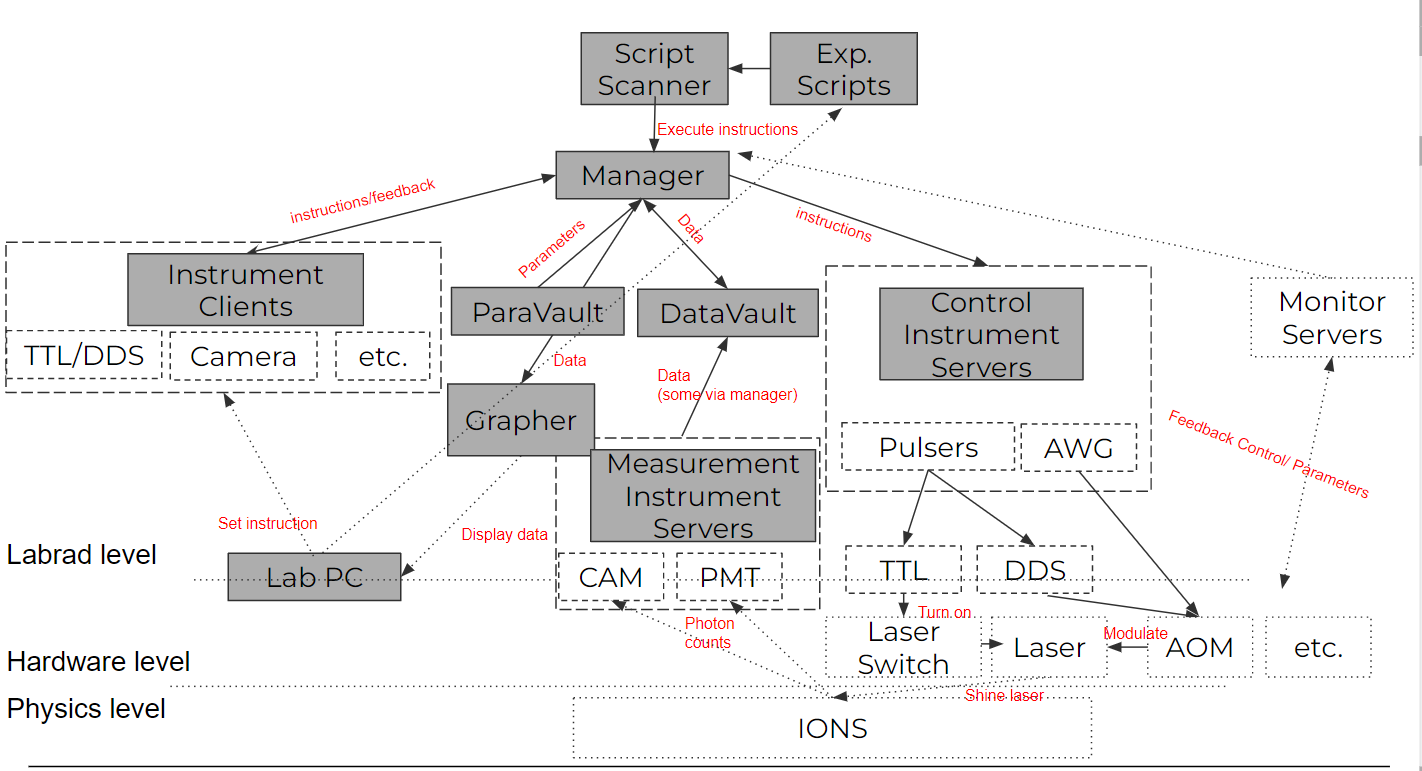
- Abstract Devices Servers: These are servers that are not associated with a specific piece of hardware, but handle tasks such as loading initial values, or running experiments, etc.(e.g. Manager, Datavault, ParameterVault, etc.)
- Labrad Manager: Talks with all servers and schedule tasks
- DataVault: Cope with all experimental data that we want to display/fit on RSG
- ParameterVault: Cope with all experimental parameters, can log/query parameters so that we can keep track of all the independent variables we fix in an experiment
- Script Scanner: A server for doing experiment properly. This script standardize the process for doing a hardware timing experiment so that you implement the experiment-related steps in an experiment file. We will focus a lot on how to use it in this section
- Control instrument Servers: These servers connected to the hardware that controls the experiment (e.g. AOM, switches) and we use them to change the independent variables we care in an experiment
- Pulser: control TTL/DDS, heart of our experiment
- AWG: control more complicated analog signals of the experiment: key to do gates, address multi-frequencies that we needed
- Measure Instrument Servers: These servers connected to the hardware that measure the experiment, they are also needed in the real time experiment
- PMT: Used to detect photons emitted by ions, no spatial degree of freedom
- Camera: Used to detect photons emitted by ions, has spatial degree of freedom
- Instrument Clients: These clients are used for displaying the instrument status/feedbacks of the experiments
- DDS/TTL Combined GUI: Used to display/control the pulser's TTL/DDS features
- PMT GUI: Used to display latest PMT data in real time
- Camera GUI(AndorVideo): Used to control and see images taken from a camera. This is not a client by itself though
- Grapher: The clients for displaying and fitting experimental data quickly after the experiment. Experimental data update in real time.
- Monitor Instrument Servers(Optional): These servers are used for monitoring the experiments' parameters. Sometimes include some feedback controls (e.g. PID) to stabilize some paramters. (e.g. wavemeters, drift trackers, Picoscope...)
Hardware timing: Revisit to the pulser sequences
FPGA vs CPU
For any quantum experiment with ions, the operations are relatively short in the microsecond ~ millisecond levels. In order to do a quantum experiment, we desperately need a precise timing mechanism. A major problem with CPU is that the clock cycles of CPUs are dynamics, meaning the timing(inverse frequency) for a CPU can change as temperature/tasks changes. Also, due to the complicated nature of jobs a CPU takes care of, codes are executed by a scheduler program, which makes the timing of operations (e.g. changing the state of a TTL channel) to be indeterministic in the millisecond level.
As a result, we need to use FPGA (in the pulser), a different kind of "CPU", to mark the timing precisely in the nanosecond level. This is also referred as the hardware timing, as in a FPGA the timing is controlled by the hardware after programming. FPGA also has a fixed clock speed around 100-1000 MHz, which makes it avoid all of the problems we discussed above. However, as we are still programming our experiment on a CPU based computer, we need to program a series of pulse sequences and send them to the FPGA so that the FPGA can execute them. For details of the programming you can check this page
Quantum Projection Noise: Repeat!
Another big thing to notice when doing quantum experiment is the quantum projection noise. As we all know, when measuring a qubit it will collapse to either a |0> or a |1> state. In fact, the quantum projection noise is kind of like an inverse problem of sampling from a binomial distribution: you need to sample N times in order to guess the p (prob of |0>) of the distribution. Therefore, in order to know the actual qubit state, we have to measure it for multiple times. So in reality we repeat the experiment and measure. This will result into a binomial distribution, and such distribution is vital when we are doing some certain experiment (e.g. calibrating the entangling gates).
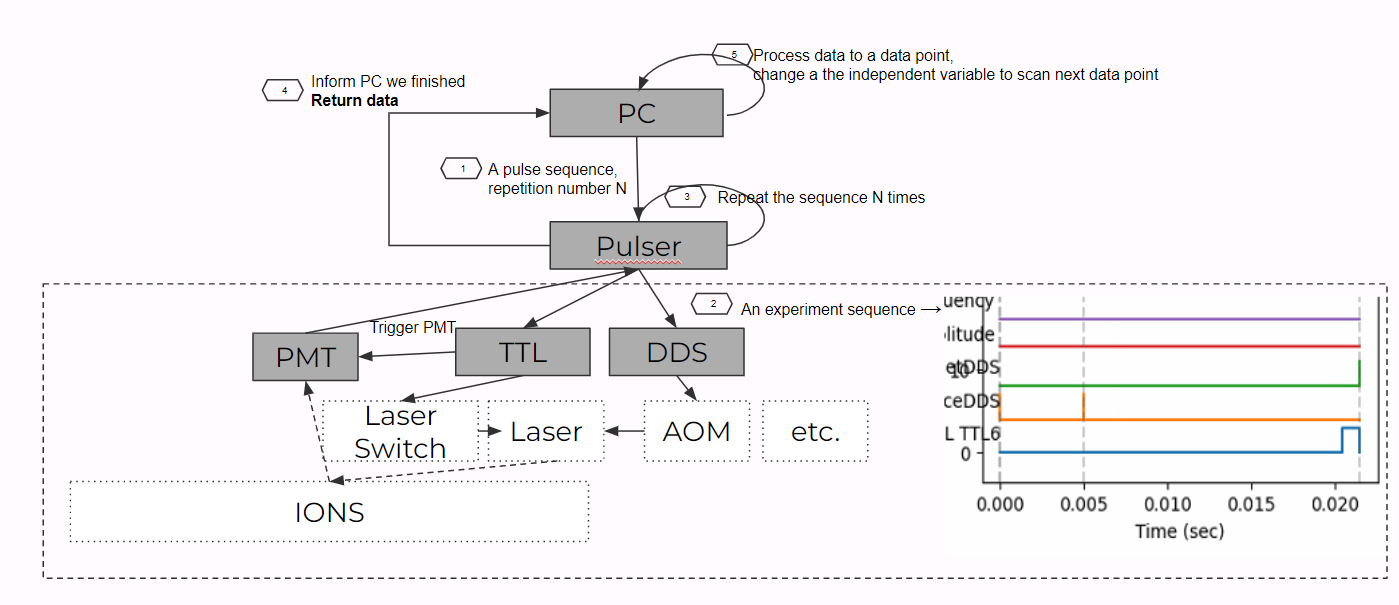
As a result, in an experiment we need to repeat for quite a lot of time. We do this by setting a constant number N (repetitions_every_point in code) so that the FPGA will repeatedly play the pulse sequence for N times. It will then notify the PC and return the data of N repetitions. The PC will then save the raw data and also process the data to make N data to a probability of p (prob of |0>) (after normalization) so that we know about the quantum states.
Scan: "Repeat" on Repeat!
When doing an experiment, we usually like to scan a list of points of a independent variable(s) and observe how the dependent variable changes. As a result, when the FPGA notify the PC that N experiments have been performed, it means that one point in the scan has been done. So then the PC will change the independent variable (usually a very similar pulses with one variable altered), and then "repeat"(or more precisely rerun with a different variable) the N experiments. The graph below shows the pipeline of what we described.
Script Scanner and experiment classes: How to run an experiment
As you can see, doing a quantum experiment is not that easy. We need quite a lot of codes for doing an experiment. This includes the instructions to the FPGAs, the data processing tools (e.g. save the raw data, averaging and normalization...), sending data to datavault and plotting them on graphers. And this can become even more complicated if you meet exceptions in an experiment. (e.g. a device is disconnected, if you want to manually end the experiment). You certainly do not want some devices to get stuck after exception as this can kills your trap or lab. So as a result, you need some codes to deal with all these things. If this is the case, we will need to write really really long scripts, which is not a good idea as debugging can be impossible. Therefore, the script scanner is implemented to help solving this. With script scanner, you only to write the parts that are different for different experiments. The script scanner is going to take care the rest.
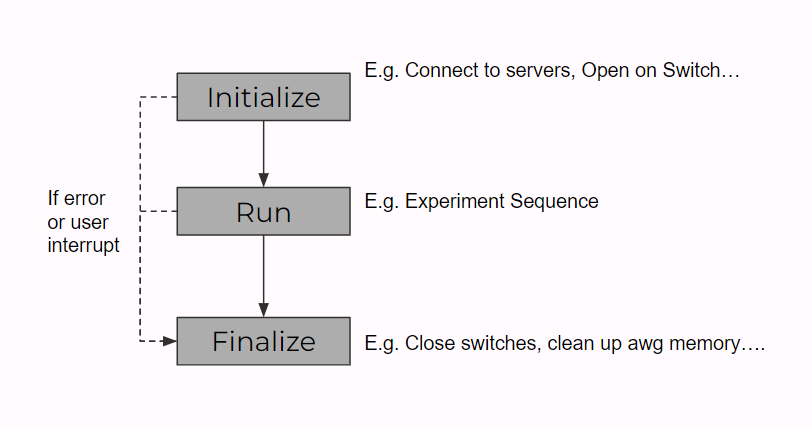
The details of the script scanner server are in the page script scanner, and in fact we haven't used many of the methods so far. So here we will talk about what methods we used in order to run an experiment. The graph below shows the structure of the experiment classes and how it interacts with the script scanner. From the bottom to top, we have sub-experiment functions (helper functions like doppler cooling sequence, calling AWG...), actual experiment classes (e.g. Scanning qubit lifetime with spin echo, Scanning frequency for a Ramsey experiment ...), scan classes (e.g. Scan_1D, Scan_1D_with_camera, Scan_2D, ...) and experiment class, experiment info class. The top two classes are fixed, it basically formats the experiment's format and how an experiment is executed (as shown by fig below below). As a result, the scan class and actual experiment both inherent from the experiment class, where we only need to implement the initialize, run, and final functions of the class to run the experiment.
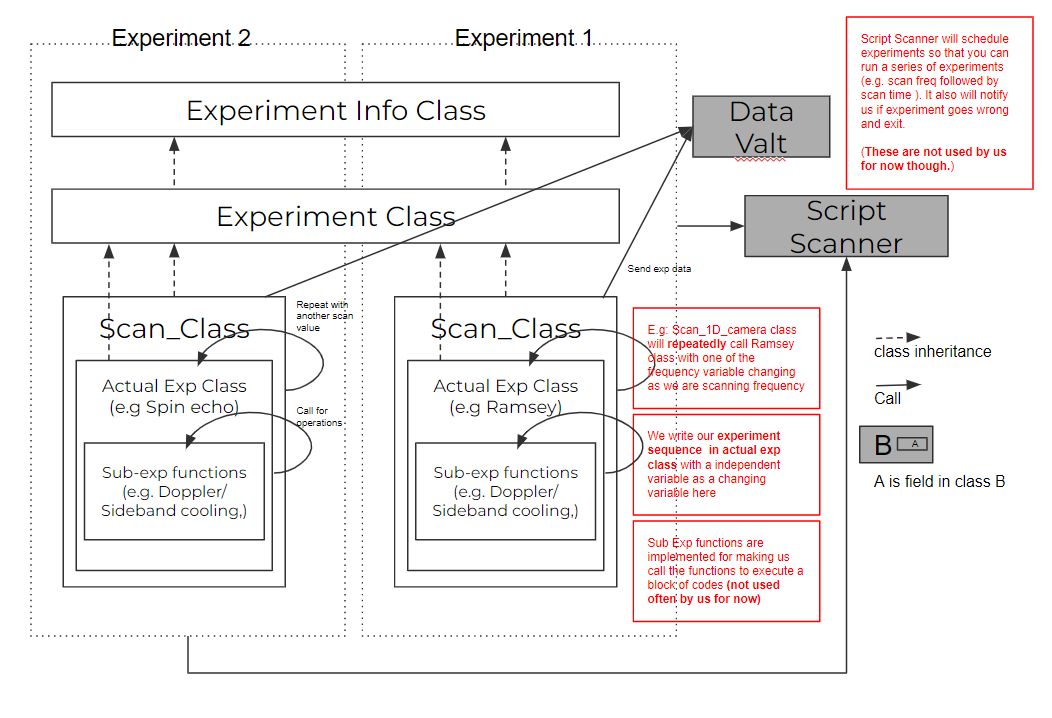
The scan classes are implemented so that we can reuse the data pipeline and structures when we scan an experiment. Basically the scan class will connect to the datavault, create a dataset of the experiment (with the name of the experiment + creation time), and run the initialization part of the actual experiment in the initialization process. It will run the actual experiment's run part with different scan values as we specified and push the data to the datavault in its own run part and will wrap up the actual experiment's finalize part and save the raw data of the experiment in its finalize part. we have variations of the scan class as we have different configurations that need different ways to build the data pipeline and execute the experiment (e.g. scan with PMT, 2D scan), but for most of the time we will stick with scan_experiment_1D_camera.py class, for which we will discuss in the section below.
With the help of the scan class, every time we want to run an experiment, we will need to implement the actual experiment class, mainly the initialize, run and finalize functions. We will open/shut down instruments in the initialize/finalize functions and implement the sequences and operations in the run functions. Note that the run functions will need to take (at least) a variable of scan value (as the scan class will pass in values) and return the experiment data (as the scan class will take it and send to the datavault). We will talk about a sample experiment in the section below too.
Examples:
Write a scan-1d experiment (with camera) based on experiment class
Here is the code for the experiment class
import traceback
import labrad
from treedict import TreeDict
from six import iteritems
from experiment_info import experiment_info
class experiment(experiment_info):
# initialization of the class
# you see we want to load cxn(command line for talking with labrad manager), pv(parameter), sc(script scanner), context(for labrad signals)
def __init__(self, name=None, required_parameters=None, cxn=None,
min_progress=0.0, max_progress=100.0,):
required_parameters = self.all_required_parameters()
super(experiment, self).__init__(name, required_parameters)
self.cxn = cxn
self.pv = None
self.sc = None
self.context = None
self.min_progress = min_progress
self.max_progress = max_progress
self.should_stop = False
# connect to cxn
def _connect(self):
if self.cxn is None:
try:
self.cxn = labrad.connect()
except Exception as error:
error_message = error + '\n' + "Not able to connect to LabRAD"
raise Exception(error_message)
try:
self.sc = self.cxn.servers['ScriptScanner']
except KeyError as error:
error_message = error + '\n' + "ScriptScanner is not running"
raise KeyError(error_message)
try:
self.pv = self.cxn.servers['ParameterVault']
except KeyError as error:
error_message = error + '\n' + "ParameterVault is not running"
raise KeyError(error_message)
try:
self.context = self.cxn.context()
except Exception as error:
error_message = error + '\n' + "self.cxn.context is not available"
raise Exception(error_message)
# excuete an experiment, you start with connecting labrad, initialize, run, finalize (implemented by subclasses)
# If we meet exception, we try to catch the message and shut down the experiment properly
def execute(self, ident):
"""Executes the experiment."""
self.ident = ident
try:
self._connect()
self._initialize(self.cxn, self.context, ident)
self._run(self.cxn, self.context)
self._finalize(self.cxn, self.context)
except Exception as e:
reason = traceback.format_exc()
print(reason)
if hasattr(self, 'sc'):
self.sc.error_finish_confirmed(self.ident, reason)
finally:
if hasattr(self, 'cxn'):
if self.cxn is not None:
self.cxn.disconnect()
self.cxn = None
def _initialize(self, cxn, context, ident):
self._load_required_parameters()
self.initialize(cxn, context, ident)
self.sc.launch_confirmed(ident)
def _run(self, cxn, context):
self.run(cxn, context)
def _load_required_parameters(self, overwrite=False):
d = self._load_parameters_dict(self.required_parameters)
self.parameters.update(d, overwrite=overwrite)
def _load_parameters_dict(self, params):
"""Loads the required parameter into a treedict."""
d = TreeDict()
for collection, parameter_name in params:
try:
value = self.pv.get_parameter(collection, parameter_name)
except Exception as e:
print(e)
message = "In {}: Parameter {} not found among Parameter Vault parameters"
raise Exception(message.format(self.name, (collection, parameter_name)))
else:
d['{0}.{1}'.format(collection, parameter_name)] = value
return d
def set_parameters(self, parameter_dict):
"""Can reload all parameter values from parameter_vault.
Alternatively, can replace parameters with values from the provided parameter_dict.
"""
if isinstance(parameter_dict, dict):
udpate_dict = TreeDict()
for (collection, parameter_name), value in iteritems(parameter_dict):
udpate_dict['{0}.{1}'.format(collection, parameter_name)] = value
elif isinstance(parameter_dict, TreeDict):
udpate_dict = parameter_dict
else:
message = "Incorrect input type for the replacement dictionary"
raise Exception(message)
self.parameters.update(udpate_dict)
def reload_some_parameters(self, params):
d = self._load_parameters_dict(params)
self.parameters.update(d)
def reload_all_parameters(self):
self._load_required_parameters(overwrite=True)
def _finalize(self, cxn, context):
self.finalize(cxn, context)
self.sc.finish_confirmed(self.ident)
# useful functions to be used in subclasses
@classmethod
def all_required_parameters(cls):
return []
def pause_or_stop(self):
"""Allows to pause and to stop the experiment."""
self.should_stop = self.sc.pause_or_stop(self.ident)
if self.should_stop:
self.sc.stop_confirmed(self.ident)
return self.should_stop
def make_experiment(self, subexprt_cls):
subexprt = subexprt_cls(cxn=self.cxn)
subexprt._connect()
subexprt._load_required_parameters()
return subexprt
def set_progress_limits(self, min_progress, max_progress):
self.min_progress = min_progress
self.max_progress = max_progress
# functions to reimplement in the subclass
def initialize(self, cxn, context, ident):
"""Implemented by the subclass."""
def run(self, cxn, context, replacement_parameters={}):
"""Implemented by the subclass."""
def finalize(self, cxn, context):
"""Implemented by the subclass."""
Here is the code for scan experiment 1d with camera class
import numpy as np
from time import localtime, strftime
from labrad.units import WithUnit
from experiment import experiment
class scan_experiment_1D_camera(experiment):
"""Used to repeat an experiment multiple times."""
# script_cls = actual experiment, we also make the list for scan values here
def __init__(self, script_cls, parameter, minim, maxim, steps, units):
self.script_cls = script_cls
self.parameter = parameter
self.units = units
self.scan_points = np.linspace(minim, maxim, steps)
self.scan_points = [WithUnit(pt, units) for pt in self.scan_points]
scan_name = self.name_format(script_cls.name)
super(scan_experiment_1D_camera, self).__init__(scan_name)
# functions for making a name for the experiment (we load the actual experiment's parameter_name )
def name_format(self, scan_name):
#Revised by Fred
return 'Scanning {0} in {1}'.format(
self.script_cls.parameter_name, scan_name)
# inherent from the initialize function of the experiment class, we run the initialize function of the actual experiment and also connect to datavault for data pipline
def initialize(self, cxn, context, ident):
self.script = self.make_experiment(self.script_cls)
self.script.initialize(cxn, context, ident)
self.navigate_data_vault(cxn, self.parameter, context)
self.script.dirc = self.dirc
# We run the actual experiment with an input of a element in the scan value, we then build the data pipeline to datavault here
# For scan with camera, we have to implement the pipeline as the image data comes back as an array
def run(self, cxn, context):
for i, scan_value in enumerate(self.scan_points):
if self.pause_or_stop():
return
#changed by Fred, only a sketchy fix, need to be fixed later
self.script.set_parameters({('para1',"para2"): scan_value})
self.script.set_progress_limits(
100.0 * i / len(self.scan_points), 100.0 * (i + 1) / len(self.scan_points))
result = self.script.run(cxn, context, scan_value)
if self.script.should_stop:
return
if result is not None:
#revised by Fred to add the feature of storing raw data
#For now raw data is handled without going to the data_vault
#When the image data is handled properly, we should also add raw data into the image class
#the logic now is that result=[plotresult,raw] for plotresult=int/float, raw=np array
#if the result=plotresult, then nothing changed
if str(type(result))=="<class 'list'>":
result[1] = np.insert(result[1],0,scan_value[self.units])
self.raw_data.append(result[1])
result = result[2]
#cxn.data_vault.add([scan_value[self.units], result], context=context)
cxn.data_vault.add(result, context=context)
self.update_progress(i)
#pass array of independent and dependent as ["name","unit"]
#Revised by Fred by adding parameter
def navigate_data_vault(self, cxn, parameter, context):
dv = cxn.data_vault
local_time = localtime()
dataset_name = self.name + strftime("%Y%b%d_%H%M_%S", local_time)
directory = ['ScriptScanner']
directory.extend([strftime("%Y%b%d", local_time), strftime("%H%M_%S", local_time)])
#need this directory to save raw_data
for text in directory:
self.dirc=self.dirc+text+'.dir/'
dv.cd(directory, True, context=context)
dv.newmatrix(dataset_name, (1,200), 'f', context=context)
dv.add_parameter('plotLive', True, context=context)
for para in parameter.keys():
dv.add_parameter(para, parameter[para], context=context)
#add parameters used in the experiment
for para in self.script.parameters:
dv.add_parameter(para,self.script.parameters[para], context=context)
#add scan points to the first element of the dataset_name
# we set 300 as this is very close to the background noise
scan_para = np.ones((1,200))*300
#print(self.scan_points)
scan_para[0,0] += self.scan_points[0][self.units]
scan_para[0,1] += self.scan_points[len(self.scan_points)-1][self.units]
scan_para[0,2] += len(self.scan_points)
dv.add(scan_para,context = context)
def update_progress(self, iteration):
progress = self.min_progress + (self.max_progress - self.min_progress) * \
float(iteration + 1.0) / len(self.scan_points)
self.sc.script_set_progress(self.ident, progress)
def finalize(self, cxn, context):
self.raw_data=np.array(self.raw_data)
#saves the raw_data into the same folder as the data vault data
if self.raw_data !=[]:
np.savetxt("../servers/data_vault/__data__/"+self.dirc+"raw_data.csv", self.raw_data, delimiter=",", fmt="%s")
self.script.finalize(cxn, context)
The code below is a script for the microwave ramsey experiment written by Fred and Aaron together
# Fred and Aaron changed the code on July 30
# We want to use awg to drive the microwave to do Ramsey experiment
# We will write detailed documentation for future reference
import labrad
from labrad.units import WithUnit
import time
import numpy as np
import sys
sys.path.append("../servers/script_scanner/")
from scan_experiment_1D_camera import scan_experiment_1D_camera
from experiment import experiment
from twisted.internet.threads import deferToThread
import msvcrt
from Andor_utils import start_camera, get_cam_status, save_analyze_imgs, start_live
#Global variables in this script:
# #of repeatitions when scanning every point
repetitions_every_point=100
# Check and change the following parameters before doing an experiment
# Choose what to scan
# TODO: Add suopport for scanning phase
scan_var = 'time'
#scan_var = 'freq'
# How far the actual transition freq comparing with the carrier transition freq we defined (at 202 MHz on AOM)
carrier_freq_offset=WithUnit(0.01-0.03,"MHz")
# Microwave pi time: this time is fixed in the experiment
microwavePiTime = WithUnit(0.0172,'ms')
# Ramsey Idle time: this time vaires when doing time scan
qubit_evolution_time = WithUnit(20,'us')
# the following parameters are used when you scan time/freq respectively
if scan_var == 'time':
# # of points you scan in an experiment
Scan_points = 100
# start time and end time
scan_time_start=WithUnit(0.0,'ms')
scan_time_end=WithUnit(1, 'ms')
# the freq you used when doing time scan
Scan_freq=WithUnit(202+ carrier_freq_offset["MHz"],'MHz')
parameter= {
'Scan_time_start':scan_time_start,
'Scan_time_end':scan_time_end,
'Scan_freq':Scan_freq,
'Scan_points':Scan_points,
'repetition at every point':repetitions_every_point,
'Raw_data_column_0':"scan freq",
"Raw_data_column_1-9":"RAW PMT Counts in 1ms with detection"
}
if scan_var == 'freq':
Scan_points = 100
Scan_freq_start = WithUnit(201.95,'MHz')
Scan_freq_end = WithUnit(202.2,'MHz')
parameter= {
'Scan_freq_start':Scan_freq_start,
'Scan_freq_end':Scan_freq_end,
'scan_time':qubit_evolution_time,
'Scan_points':Scan_points,
'repetition at every point':repetitions_every_point,
'Raw_data_column_0':"scan freq",
"Raw_data_column_1-9":"RAW PMT Counts in 1ms with detection"
}
#Define all experiment parameters, program as {('name','labrad unit'),....}
#scripts for the scanning part. For now we will send a trigger pulser of 1ms from the TTL1 chanel of the pulser once it is runned
class scan(experiment):
name = 'MicrowaveRamseyScan'
parameter_name = "DDS Freq"
required_parameters = [('cooling', 'detection_freq'),
('cooling', 'detection_pwr'),
('cooling', 'detection_time'),
('cooling', 'doppler_cooling_pwr'),
('cooling', 'doppler_cooling_time'),
('cooling', 'far_detuned_cooling_pwr'),
('cooling', 'far_detuned_cooling_time'),
('cooling', 'far_detuned_pumping_pwr'),
('cooling', 'optical_pumping_time'),
('cooling', 'repumper_pumping_pwr'),
('cooling', 'repumper_cooling_pwr'),
('cooling', 'DDS_delay'),
]
USEPMT = False
# parameters for camera
cam_var = {
'identify_exposure': WithUnit(1, 'ms'),
'start_x' : 896,
'stop_x' : 1095,
'start_y' : 68,
'stop_y' : 83,
'horizontalBinning' : 1,
'verticalBinning' : 16,
'kinetic_number' : repetitions_every_point,
'counter': 0,
'USEPMT' : USEPMT
}
#Note the following function is used to load parameters from pv
#This is needed for all experiments that needs to use parameter vault
@classmethod
def all_required_parameters(cls):
parameters = set(cls.required_parameters )
parameters = list(parameters)
return parameters
#you program the experimental sequence here for DDS and TTL
#the logic here is to pack all the TTL/DDS command sequences of scanning 1 point together and send to FPGA on pulser
#unit in ms, you will need to change the time to the correct para if you scan freq and vice versa
def program_main_sequence(self, scanfreq, scantime):
#We set up these values in the registry
far_detuned = self.parameters.cooling.far_detuned_cooling_time
doppler = self.parameters.cooling.doppler_cooling_time
detection = self.parameters.cooling.detection_time
optical_pumping_time = self.parameters.cooling.optical_pumping_time
repumper_pumping_pwr = self.parameters.cooling.repumper_pumping_pwr
far_detuned_pumping_pwr = self.parameters.cooling.far_detuned_pumping_pwr
doppler_cooling_pwr = self.parameters.cooling.doppler_cooling_pwr
detection_pwr = WithUnit(-11,"dBm")
repumper_cooling_pwr = self.parameters.cooling.repumper_cooling_pwr
far_detuned_cooling_pwr = self.parameters.cooling.far_detuned_cooling_pwr
#repumping freqs are calculated using detection freq
detection_freq = WithUnit(139.5,"MHz")
repumper_pumping_freq = detection_freq*2 - WithUnit(120.6-1,"MHz")
far_detuned_puming_freq = WithUnit(270.7+1,"MHz") -detection_freq
doppler_cooling_freq = detection_freq - WithUnit(4.5, 'MHz')
DDS_delay = self.parameters.cooling.DDS_delay
# in the original function scantime is a unitless values
# we make scantime a value with unit
scantime = WithUnit(scantime,"ms") + DDS_delay
#we define the following times for keeping track of different state of the experiment
cooling_stop_at = far_detuned+WithUnit((0.4), 'ms')+WithUnit(0.1, 'ms')
# we add 10us to wait
optical_pumping_stop_at = cooling_stop_at + optical_pumping_time + WithUnit(45, 'us')
second_MW_drive_start_at = optical_pumping_stop_at+microwavePiTime/2+scantime
#we add 100 us wait time (if we use raman we need to wait much longer)
camera_start_at = second_MW_drive_start_at + microwavePiTime/2+ WithUnit(2000, 'us')
#this is always needed if you want to start a new sequence
self.pulser.new_sequence()
# MW pi/2 we minus 500ns for the TTL to open AWG
self.pulser.add_ttl_pulse('TTL3', optical_pumping_stop_at-WithUnit(0.0005, 'ms') , microwavePiTime/2)
# MW pi pulse for echo
#self.pulser.add_ttl_pulse('TTL3', far_detuned+WithUnit(0.55-0.0005, 'ms')+scantime/2, WithUnit(microwavePiTime,'ms'))
# MW pi/2
self.pulser.add_ttl_pulse('TTL3', second_MW_drive_start_at-WithUnit(0.0005, 'ms'), microwavePiTime/2)
#Camera Trigger
self.pulser.add_ttl_pulse('TTL6', camera_start_at, detection)
self.pulser.add_ttl_pulse('TTL2', camera_start_at, detection)
#extend sequence
self.pulser.extend_sequence_length(WithUnit(7.5, 'ms'))
#you add DDS pluse by the following code: [(DDS pulse 1),(DDS pulse 2),..]
#for DDS pulse, you program by (name,start_time,duration,freq,power,phase,ramp freq, ramp pow)
DDS = [
#far_detuned/repumper Cooling
('DDS7', WithUnit(0.001, 'ms'), far_detuned, repumper_pumping_freq, repumper_cooling_pwr, WithUnit(0.0, 'deg'), WithUnit(0, 'MHz'),WithUnit(0, 'dB')),
('DDS8', WithUnit(0.001, 'ms'), far_detuned, far_detuned_puming_freq, far_detuned_cooling_pwr, WithUnit(0.0, 'deg'), WithUnit(0, 'MHz'),WithUnit(0, 'dB')),
#Doppler Cooling
('DDS6', -doppler+far_detuned+WithUnit((0.2), 'ms'), doppler, doppler_cooling_freq, doppler_cooling_pwr, WithUnit(0.0, 'deg'), WithUnit(0, 'MHz'),WithUnit(0, 'dB')),
('DDS6', far_detuned+WithUnit((0.25), 'ms'), WithUnit(0.05, 'ms'), doppler_cooling_freq, doppler_cooling_pwr + WithUnit(-2.0, 'dBm'), WithUnit(0.0, 'deg'), WithUnit(0, 'MHz'),WithUnit(0, 'dB')),
('DDS6', far_detuned+WithUnit((0.3), 'ms'), WithUnit(0.1, 'ms'), doppler_cooling_freq, doppler_cooling_pwr + WithUnit(-4.0, 'dBm'), WithUnit(0.0, 'deg'), WithUnit(0, 'MHz'),WithUnit(0, 'dB')),
('DDS6', far_detuned+WithUnit((0.4), 'ms'), WithUnit(0.1, 'ms'), doppler_cooling_freq, doppler_cooling_pwr + WithUnit(-6.0, 'dBm'), WithUnit(0.0, 'deg'), WithUnit(0, 'MHz'),WithUnit(0, 'dB')),
#Optical pumping
('DDS7', cooling_stop_at, optical_pumping_time, repumper_pumping_freq, repumper_pumping_pwr, WithUnit(0.0, 'deg'), WithUnit(0, 'MHz'),WithUnit(0, 'dB')),
('DDS8', cooling_stop_at, optical_pumping_time, far_detuned_puming_freq, far_detuned_pumping_pwr, WithUnit(0.0, 'deg'), WithUnit(0, 'MHz'),WithUnit(0, 'dB')),
#Microwave is here, but we will use TTL3 to trigger the window in this time period
#detection
('DDS6', camera_start_at, detection, detection_freq, detection_pwr, WithUnit(0.0, 'deg'), WithUnit(0, 'MHz'),WithUnit(0, 'dB'))
]
# program DDS
self.pulser.add_dds_pulses(DDS)
# you need this to compile the acutal sequence you programmed
self.pulser.program_sequence()
def initialize(self, cxn, context, ident):
try:
self.pulser = self.cxn.pulser
#self.pmt = self.cxn.normalpmtflow
self.awg = self.cxn.keysight_awg
self.cam = self.cxn.andor_server
self.dv = self.cxn.data_vault
except Exception as e:
print(e)
#refresh pv
self.pv = self.cxn.parametervault
self.pv.reload_parameters()
# Initialize AWG for microwave
# note awg chanel 3 is tied with microwave
self.awg.amplitude(3,WithUnit(1.49,"V"))
self.pulser.switch_manual('TTL3',False)
self.pulser.switch_auto('TTL3')
print('init, set up pulser')
def run(self, cxn, context, scanvalue):
# this is used for stop the experiment with esc key
if msvcrt.kbhit():
if ord(msvcrt.getch()) == 27:
print('exit the experiment')
self.should_stop = True
print('scanning at '+str(scanvalue))
#you can program the sequence in the program_main_sequence function
if scan_var == 'freq':
#we need to reconfigure AWG here:
#print(scanvalue)
self.awg.frequency(3,scanvalue)
self.program_main_sequence(scanfreq = scanvalue['MHz'],scantime=qubit_evolution_time['ms'])
elif scan_var == 'time':
#we need to reconfigure AWG here:
self.awg.frequency(3,Scan_freq)
self.program_main_sequence(scanfreq= Scan_freq['MHz'], scantime = scanvalue['ms'])
#you set TTL channels you needed to be auto to control them
self.pulser.switch_auto('TTL3')
self.pulser.switch_auto('TTL2')
self.pulser.switch_auto('TTL6')
#config camera
start_camera(self.cam, self.cam_var)
#you play the sequence for n times (n=100 here), i.e. number of repeatitions per scan point.
self.pulser.start_number(repetitions_every_point)
#you wait the sequence to be done
self.pulser.wait_sequence_done()
self.pulser.stop_sequence()
# get results: PMT or Camera
if self.USEPMT ==True:
readout = self.pulser.get_readout_counts()
count = np.array(readout)#[readout>15]
avg_count = np.average(count)
get_cam_status(self.cam)
save_analyze_imgs(self.cam, self.cam_var, self.dirc)
avg_image = []
else:
start_time = time.time()
get_cam_status(self.cam)
print("wait cam--- %s seconds ---" % (time.time() - start_time))
start_time = time.time()
readout,avg_image = save_analyze_imgs(self.cam, self.cam_var, self.dirc)
#print(readout)
count = np.array(readout)#[readout>15]
avg_count = np.average(count)
print("process data--- %s seconds ---" % (time.time() - start_time))
#you switch back the TTL channels into manual
self.pulser.switch_manual('TTL3')
self.pulser.switch_manual('TTL2')
self.pulser.switch_manual('TTL6')
return [avg_count,readout, avg_image]
def finalize(self, cxn, context):
print('finalize')
# Here we start to really run this experiment
if __name__ == '__main__':
#connect to cxn
cxn = labrad.connect()
#connect to script scanner
scanner = cxn.scriptscanner
###: you set the scan freqeuncy and data points needed here
# we call the scan_experiment_1D_camera class with all the experiment parameters and the entire scan class (which is the actual experiment's class above)
if scan_var=='freq':
exprt = scan_experiment_1D_camera(scan, parameter, Scan_freq_start['MHz'], Scan_freq_end['MHz'], Scan_points, 'MHz')
elif scan_var=='time':
exprt = scan_experiment_1D_camera(scan, parameter, scan_time_start['ms'], scan_time_end['ms'], Scan_points, 'ms')
# Now as we have made the class of scan_experiment_1D_camera with our actual experiment class (scan class, forgive the bad naming here)
# We use the register_external_launch function to let the script scanner to add 1 script to the queue (nothing in the queue though)
ident = scanner.register_external_launch(exprt.name)
# We now execute the experiment (this function is in the experiment class), and we use script scanner to log the error if there are any
exprt.execute(ident)