Project 2 - ncbi/workshop-asm-ngs-2022 GitHub Wiki
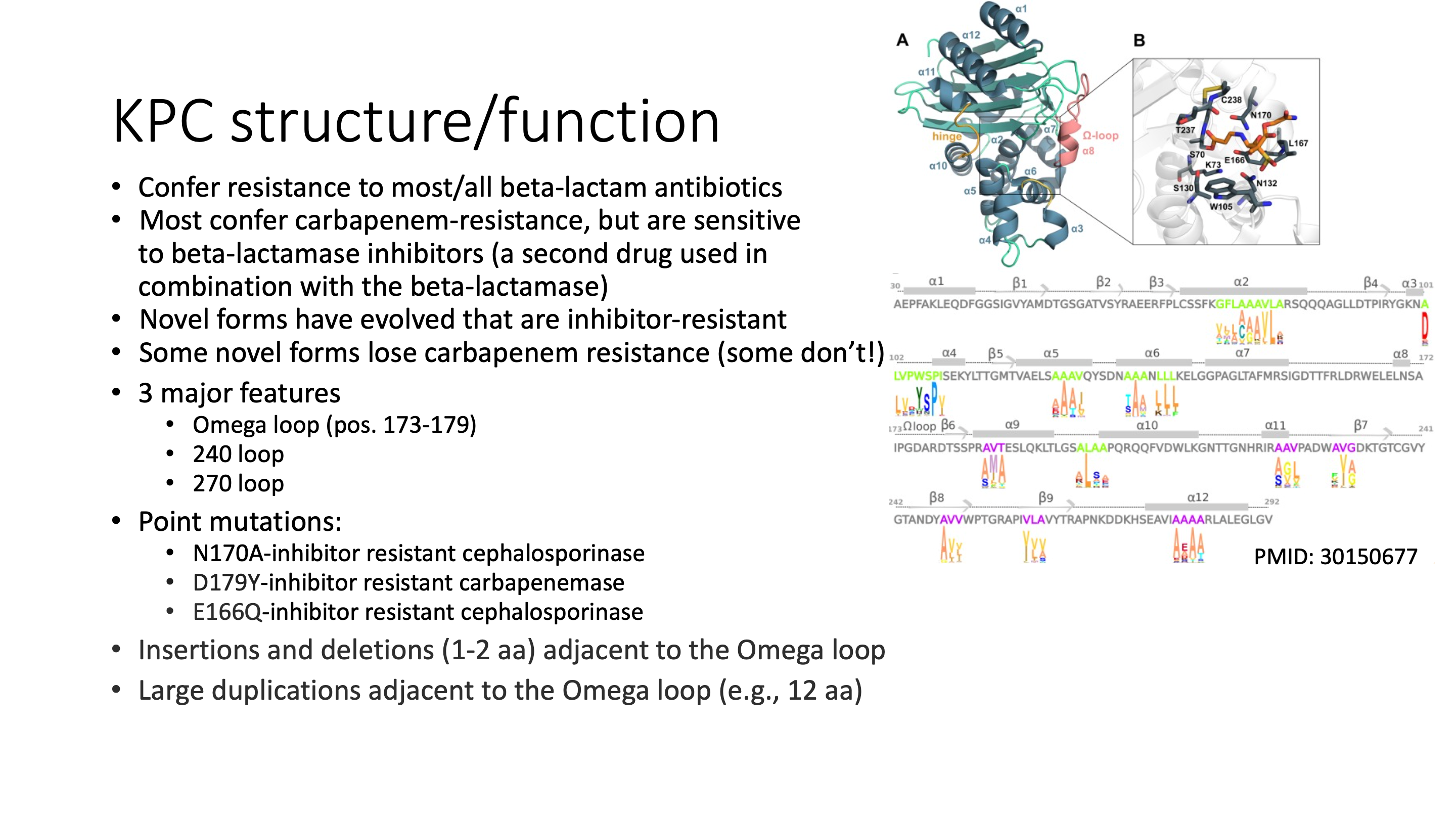
blaKPC genes are beta-lactamases that can hydrolize cabapenems, which are last-line beta-lactam drugs. Though very closely related there are functional differences in substrate specificity and avibactam resistance.
Instructions on setting up a VM for this and Project 2 are in Setup.
For this project we're going to be working in the web console we already opened. If you have closed it you need to click the 'SSH' link next to the listing for your VM in the Compute Engine VM instances list.
mkdir -p ~/project2
cd ~/project2The AMRFinderPlus database contains a specially formatted FASTA file that includes the coding sequence for all the genes in the Reference Gene Catalog. You can also download the coding sequence from the Reference Gene Catalog web interface.
wget https://ftp.ncbi.nlm.nih.gov/pathogen/Antimicrobial_resistance/AMRFinderPlus/database/latest/AMR_CDSThere are a few KPC alleles with large insertions that we will exclude in this analysis to simplify demonstrationa and analysis.
The data behind the Reference Gene Catalog is available on our FTP site at https://ftp.ncbi.nlm.nih.gov/pathogen/Antimicrobial_resistance/AMRFinderPlus/database/latest/ReferenceGeneCatalog.txt. We have documentation for the files in the AMRFinderPlus database on the AMRFinderPlus wiki
wget https://ftp.ncbi.nlm.nih.gov/pathogen/Antimicrobial_resistance/AMRFinderPlus/database/latest/ReferenceGeneCatalog.txtWe'll use awk to calculate the length of each element in nucleotide space and filter for those less than 297 amino acids (genbank_stop - genbank_start + 1 < 297). For those lines that match we'll print out the refseq_accession, allele, and the length in amino-acids.
awk -F'\t' '$2 == "blaKPC" && ($20-$19+1)/3 < 297 { print $11"\t"$1"\t"($20-$19+1)/3-1 }' ReferenceGeneCatalog.txt > kpc_to_analyze.tabkpc_to_analyze.tab is in the format accession <tab> allele <tab> length
Filter the FASTA file for the sequences we're interested in and make convenient phylip-compatible names for visualizing on a phylogenetic tree.
time while read -u 10 accession allele length
do
seqkit grep -r -p "$accession" AMR_CDS |
sed "s/^>.*/>${accession}_${allele}_${length}/"
done 10< kpc_to_analyze.tab > kpc_to_analyze.fnamuscle -align kpc_to_analyze.fna -output kpc_to_analyze.aln.fnaThis step should take a couple of minutes.
muscle -align kpc_to_analyze.fna -output kpc_to_analyze.aln.fna
muscle 5.1.linux64 [12f0e2] 32.9Gb RAM, 8 cores
Built Jan 13 2022 23:17:13
(C) Copyright 2004-2021 Robert C. Edgar.
https://drive5.com
Input: 100 seqs, avg length 882, max 888
00:00 4.6Mb CPU has 8 cores, running 8 threads
01:57 541Mb 100.0% Calc posteriors
01:59 554Mb 100.0% Consistency (1/2)
02:01 554Mb 100.0% Consistency (2/2)
02:01 554Mb 100.0% UPGMA5
02:03 565Mb 100.0% Refining
raxml-ng --search --msa kpc_to_analyze.aln.fna --model GTR+I+G --seed 1 --redoThe end of the output from RAxML-NG run should look something like this:
Best ML tree with collapsed near-zero branches saved to: /home/aprasad/project2/kpc_to_analyze.aln.fna.raxml.bestTreeCollapsed
Best ML tree saved to: /home/aprasad/project2/kpc_to_analyze.aln.fna.raxml.bestTree
All ML trees saved to: /home/aprasad/project2/kpc_to_analyze.aln.fna.raxml.mlTrees
Optimized model saved to: /home/aprasad/project2/kpc_to_analyze.aln.fna.raxml.bestModel
Execution log saved to: /home/aprasad/project2/kpc_to_analyze.aln.fna.raxml.log
Analysis started: 21-Sep-2022 18:02:17 / finished: 21-Sep-2022 18:02:56
Elapsed time: 39.329 seconds
We're using the web-based software iTOL for this exercise, an alternative to using iTOL to visualize the tree is NCBI's desktop GUI software Genome Workbench.
cat kpc_to_analyze.aln.fna.raxml.bestTreeCopy the text of the tree.
Your tree should look like, if you don't use or change the --seed option above the tree might vary because of differences in the random numbers used by RAxML-NG:
((((NG_073466.1_blaKPC-63_293:0.001298,NG_049261.1_blaKPC-7_293:0.001286):0.000001,(((NG_076667.1_blaKPC-91_293:0.000001,NG_079893.1_blaKPC-118_293:0.001293):0.001334,(((NG_081699.1_blaKPC-130_293:0.001298,(NG_071204.1_blaKPC-78_293:0.001311,NG_080778.1_blaKPC-125_293:0.000001):0.001309):0.000001,(NG_049252.1_blaKPC-19_293:0.002706,(NG_065878.1_blaKPC-46_293:0.001300,(((NG_073465.1_blaKPC-62_293:0.001304,(NG_079890.1_blaKPC-115_291:0.002847,NG_079233.1_blaKPC-92_291:0.000001):0.001353):0.000001,NG_076680.1_blaKPC-94_292:0.000001):0.000001,NG_070739.1_blaKPC-66_291:0.000001):0.000001):0.000001):0.000001):0.000001,(NG_078032.1_blaKPC-98_293:0.001285,(NG_049254.1_blaKPC-21_293:0.001310,NG_052862.1_blaKPC-27_293:0.000001):0.001300):0.000001):0.000001):0.000001,NG_049257.1_blaKPC-3_293:0.000001):0.000001):0.000001,((((NG_074718.1_blaKPC-69_295:0.001284,(((NG_074715.1_blaKPC-48_293:0.001289,NG_074714.1_blaKPC-47_293:0.001287):0.000001,NG_063841.1_blaKPC-39_293:0.000001):0.000001,NG_081071.1_blaKPC-127_295:0.001282):0.001277):0.000001,(NG_064726.1_blaKPC-40_295:0.001286,NG_052581.1_blaKPC-28_291:0.000001):0.000001):0.000001,(NG_068176.1_blaKPC-53_295:0.000001,(((NG_079897.1_blaKPC-122_293:0.001285,NG_070180.1_blaKPC-61_293:0.001296):0.000001,NG_049246.1_blaKPC-13_293:0.002597):0.000001,(NG_068016.1_blaKPC-56_293:0.001286,NG_071203.1_blaKPC-49_293:0.001295):0.000001):0.000001):0.000001):0.000001,((NG_076666.1_blaKPC-90_295:0.000001,(NG_051167.1_blaKPC-25_295:0.000001,(NG_079894.1_blaKPC-119_293:0.001310,(NG_068177.1_blaKPC-55_293:0.001325,(NG_079231.1_blaKPC-83_293:0.001311,(((((((((((NG_070743.1_blaKPC-75_293:0.001308,(((((((((NG_073471.1_blaKPC-82_295:0.000001,(NG_079891.1_blaKPC-116_293:0.001308,((((NG_065877.1_blaKPC-45_293:0.001320,((((((NG_080779.1_blaKPC-126_293:0.000001,NG_074721.1_blaKPC-85_293:0.001284):0.000001,NG_070740.1_blaKPC-72_293:0.001311):0.001257,(NG_070895.1_blaKPC-71_294:0.000001,((NG_073467.1_blaKPC-64_294:0.002546,(NG_079896.1_blaKPC-121_294:0.000001,NG_074717.1_blaKPC-68_295:0.000001):0.000001):0.001225,NG_079889.1_blaKPC-114_295:0.000001):0.000001):0.001879):0.001985,NG_067226.1_blaKPC-54_293:0.001310):0.000001,NG_064728.1_blaKPC-43_293:0.001318):0.000001,NG_064727.1_blaKPC-42_293:0.001318):0.000001):0.000001,NG_074723.1_blaKPC-87_292:0.000001):0.000001,NG_049251.1_blaKPC-18_293:0.001310):0.000001,(((NG_049255.1_blaKPC-22_293:0.001336,NG_049249.1_blaKPC-16_293:0.002643):0.000001,NG_049250.1_blaKPC-17_293:0.000001):0.000001,NG_061612.1_blaKPC-37_293:0.001329):0.001318):0.000001):0.000001):0.000001,(NG_054685.1_blaKPC-30_293:0.000001,NG_049256.1_blaKPC-24_293:0.001319):0.001309):0.000001,NG_073470.1_blaKPC-81_292:0.000001):0.000001,NG_081700.1_blaKPC-131_293:0.001318):0.000001,NG_070178.1_blaKPC-59_293:0.001311):0.000001,NG_074720.1_blaKPC-84_293:0.001335):0.000001,NG_070897.1_blaKPC-77_293:0.001380):0.000001,(NG_074722.1_blaKPC-86_293:0.001028,NG_068508.1_blaKPC-57_293:0.001088):0.000518):0.000001,NG_051469.1_blaKPC-26_293:0.001310):0.000001):0.000001,(NG_079230.1_blaKPC-112_289:0.000001,(NG_049247.1_blaKPC-14_291:0.000001,(NG_079232.1_blaKPC-89_293:0.001285,NG_081070.1_blaKPC-100_293:0.000001):0.001303):0.000001):0.000001):0.000001,((NG_049243.1_blaKPC-10_293:0.001307,(((NG_049248.1_blaKPC-15_293:0.009645,NG_049258.1_blaKPC-4_293:0.000001):0.001337,NG_079895.1_blaKPC-120_293:0.001313):0.000001,NG_049259.1_blaKPC-5_293:0.000001):0.000001):0.001314,NG_049244.1_blaKPC-11_293:0.000001):0.001305):0.000001,NG_074724.1_blaKPC-88_293:0.001310):0.000001,((NG_049260.1_blaKPC-6_293:0.000001,NG_049262.1_blaKPC-8_293:0.001326):0.001332,NG_070742.1_blaKPC-74_291:0.000001):0.000001):0.000001,NG_060524.1_blaKPC-35_293:0.001326):0.000001,NG_049245.1_blaKPC-12_293:0.001320):0.000001,NG_049253.1_blaKPC-2_293:0.000001):0.000001,NG_070179.1_blaKPC-60_293:0.001310):0.000001,NG_079888.1_blaKPC-113_294:0.000001):0.000001,NG_078037.1_blaKPC-96_293:0.001330):0.000001):0.000001):0.000001):0.000001):0.000001):0.001211,NG_073468.1_blaKPC-65_295:0.000001):0.000001):0.000001):0.000001,((NG_067224.1_blaKPC-51_293:0.003867,((NG_055494.1_blaKPC-31_293:0.000001,(NG_076681.1_blaKPC-95_293:0.001259,((NG_056170.1_blaKPC-33_293:0.000001,NG_067225.1_blaKPC-52_294:0.000001):0.000001,NG_078063.1_blaKPC-102_293:0.001304):0.001275):0.000001):0.000001,(NG_074719.1_blaKPC-70_293:0.001267,(NG_055495.1_blaKPC-32_293:0.000001,NG_081072.1_blaKPC-128_293:0.001258):0.001255):0.000001):0.000954):0.000458,NG_062357.1_blaKPC-38_293:0.001293):0.000001,(NG_060569.1_blaKPC-23_293:0.001299,NG_061389.1_blaKPC-36_293:0.001295):0.000001);
Go go iTOL at https://itol.embl.de/upload.cgi and copy and paste your tree into the upload box.
You can see your tree, but we'll add an annotation file to color branches so we can see the length variation more easily.
There are two annotation files we used to add color and phenotype information to the tree at iTOL. Documentation for the annotation file formats at iTOL is here.
Here are the two annotation files we've already generated. Right click each of them and save them for upload to iTOL.
To save time we've run them already and link to them here, but the following commands will generate them live from your data. You can also take a look at the two quick-and-dirty scripts to generate the annotation files: make_tree_colors.pl and annotate_phenotypes.pl.
wget https://raw.githubusercontent.com/wiki/ncbi/workshop-asm-ngs-2022/make_tree_colors.pl
perl make_tree_colors.pl kpc_to_analyze.tab > tree_colors.txt
wget https://raw.githubusercontent.com/wiki/ncbi/workshop-asm-ngs-2022/annotate_phenotypes.pl
perl annotate_phenotypes.pl ReferenceGeneCatalog.txt > phenotype_annotation.txtClick the "Datasets" tab in iTOL and then the "Upload annotation files" button.
Select the tree_colors.txt and phenotype_annotation.txt files you already downloaded and click upload.
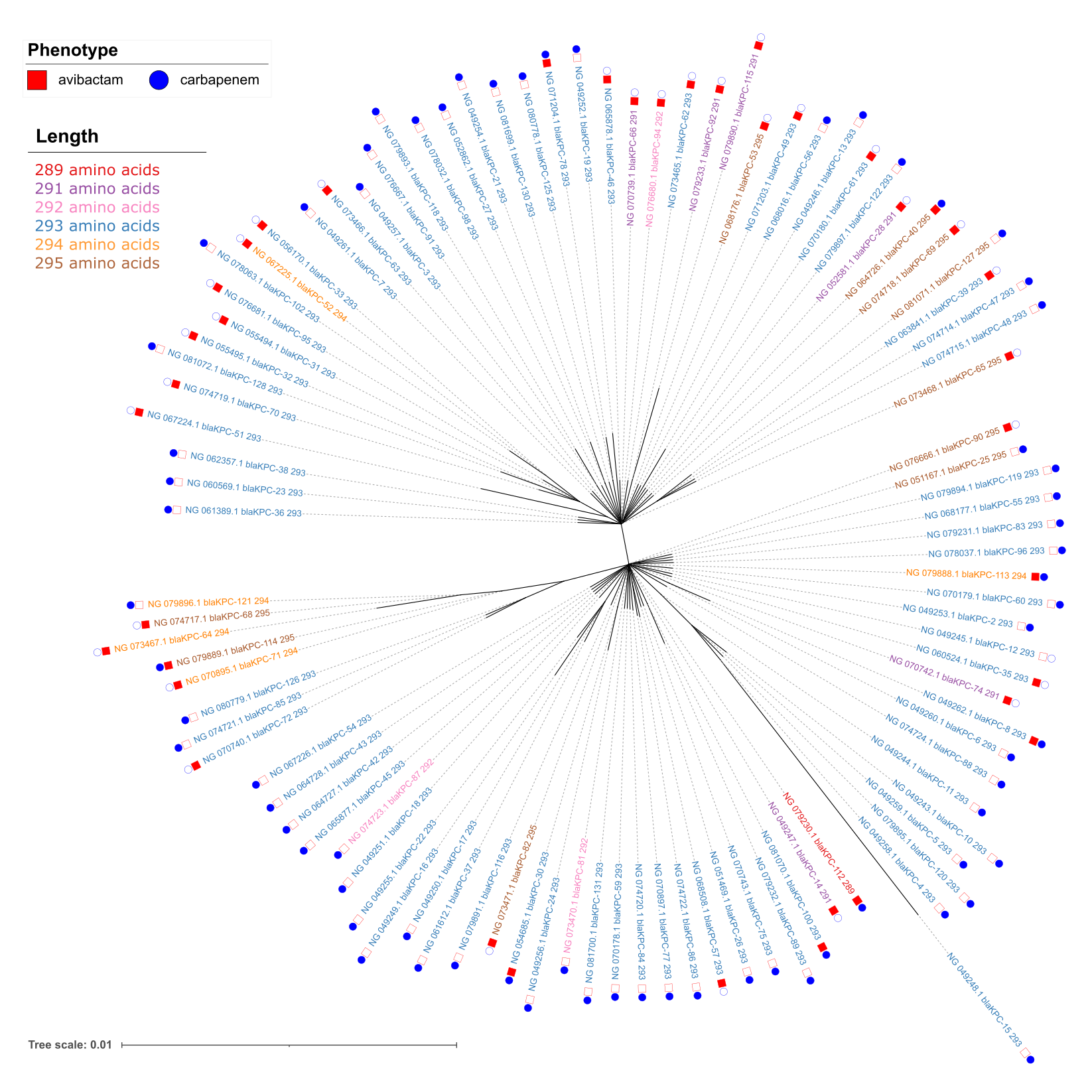
Three conclusions:
- "Star topology" indicates a recent radiation from ancestor sequence with little resolution in the tree. Little data to verify exact branching pattern.
- Likely multiple independent mutational events leading to size differences.
- Some phenotypes have not been assessed experimentally and that could be a source of inconsistency.
Continue on to Project 3