Miniproject: Activity - petermr/CEVOpen GitHub Wiki
- Development of activity dictionary to serve as a tool searching and annotating scientific articles
- Testing of getpapers; a web scrapper for open-source scientific literature and using it for the creation of a corpus of medicinal activity and essential oils scientific papers
- Running a dictionary-based search within the created corpus and drawing relationships between plant sources and compounds with medicinal activity
- Creating Dictionary using
getpapersandamiquery. Installation of getpapers & ami - Medicinal plants have been used for many years for therapy and prevention of various human diseases because they have always shown many different biological activities (e.g., antimicrobial, antioxidant, anti-inflammatory, anti-cancer).
-
getpapersis a simple, powerful tool for querying repositories of scholarly articles using a simple one-line command. - It collects all freely available research papers in full text and xml format to your local machine.
- The command getpapers will initiate the process and -q refers to the query which is to be searched. The query is entered in inverted commas as is done in "(medicinal activity) AND (essential oil)". The next element is -o which refers to output directory and the parameter that follows it in the name of the directory which is eo_activity in our case. Then, -x -p corresponds to xml and pdf files to be included in our search and -k 100 limits our search to 100 files only.
- getpapers used to create corpus of medicinal activities
General code syntax: getpapers -q <"project title "> -o <file name> -x<xml> -p<pdf> -k <number of papers requied>
Query code:
getpapers -q "(medicinal activity) AND (essential oil)" -o eo_activity -x -p -k 100 -f activity/log.txt
This helps to build corpus of 92 articles with full text and xml file
- ami is a framework for gathering, searching, transforming scholarly publications, oriented towards STEMM (Science, technology, Engineering, Medicine, Maths).
Ami section which is used to section the research papers into the front, body, back ,floats and groups. Sectioning of downloaded files will create a tree structure for us which will help in exploring the content of the file. Sectioning done using section function of ami .Which runs on command prompt.
General code syntax: ami -p <cproject> section
Query code:
ami –p eo_activity section
Ami search which search and analysis the terms in your project repository and gives the frequency is terms and the histogram of your corpus.
General code syntax: ami –p <cprooject><directory> search –dictionary <path>
Query code:
ami -p eo_activity search --dictionary activity
For search_lib, download PAPERS into PROJECTS , find SECTIONS and index with (DICTIONARIES and/or PATTERNS) into a searchable KNOWLEDGEBASE and analyse for new INSIGHTS.
General code syntax: python search_lib.py --dict --sect --proj
Query code:
python search_lib.py --dict activity --sect introduction --proj activity
python search_lib.py --dict activity --sect METHOD --proj oil186
Collected freely available papers from EUROPMC. Once getpapers command executive.
FIGURE 1: OUTPUT OF getpapers
Results of ami section. It sections the papers in the directory.
FIGURE 2: OUTPUT OF ami section
Results are in the form of table , histogram and in the each folder results.

FIGURE 4: OUTPUT OF AMI SEARCH IN TABLE WITH FREQUENCY
FIGURE 5: PLOT OF .SVG FILE
Results are in the form of matplotlib graph for activity dictionary with corpus activity and oil186
FIGURE 6: OUTPUT OF SEARCH_LIB WITH SECTION INTRODUCTION AND MINICORPUS ACTIVITY
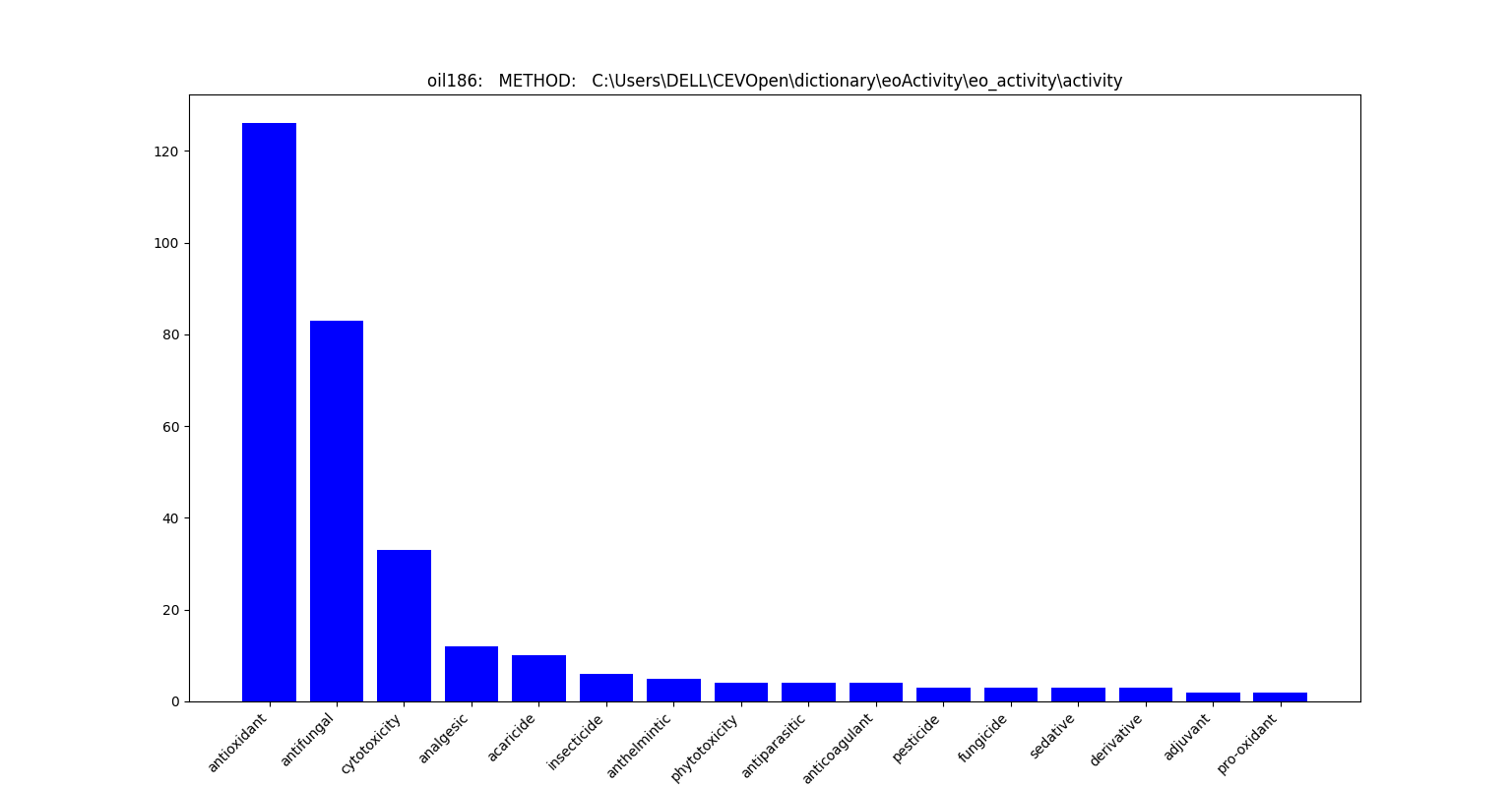
FIGURE 7: OUTPUT OF SEARCH_LIB WITH SECTION METHOD AND MINICORPUS OIL186