Pacific Biosciences: RSII - aechchiki/SIB_LongReadsWorkshop_Zurich17 GitHub Wiki
The instrument
The RSII is a sequencing instrument providing long reads, and commercialized by PacBio:

The sequencing technology
Here you can find a video briefly explaining the technology.
Sequencing calls on the PacBio RSII platform are based on the optical detection of the incorporation of phospholinked nucleotide, in form of a movie.
This is the principle behind the SMRT (Single Molecule Real Time) sequencing, and happens in tiny ZMWs microwells (Zero Mode Waveguides) directly on the sequencing flowcell (SMRTcell):

At the bottom of each ZMWs, a natural polymerase incorporates complementary bases to a DNA/cDNA fragment:

The polymerase continuously 'reads' the fragment to be sequenced, which has to be prepared with hairpin adapters (SMRTbell), where the polymerase can bind to start reading the template. SMRTbells are ligated on both sides of each double-stranded DNA fragment, forming a circular sequence:

The fragments to be sequenced can differ in length. This implies that, for a given sequencing time, short DNA fragments will be read many times, while long fragments may be read just few times completely.
The complete sequence is called a polymerase read. After the adapter sequences are removed, the sequence is split into subreads. The consensus of subreads is called a circular consensus read (CCS) or read of insert (ROI).

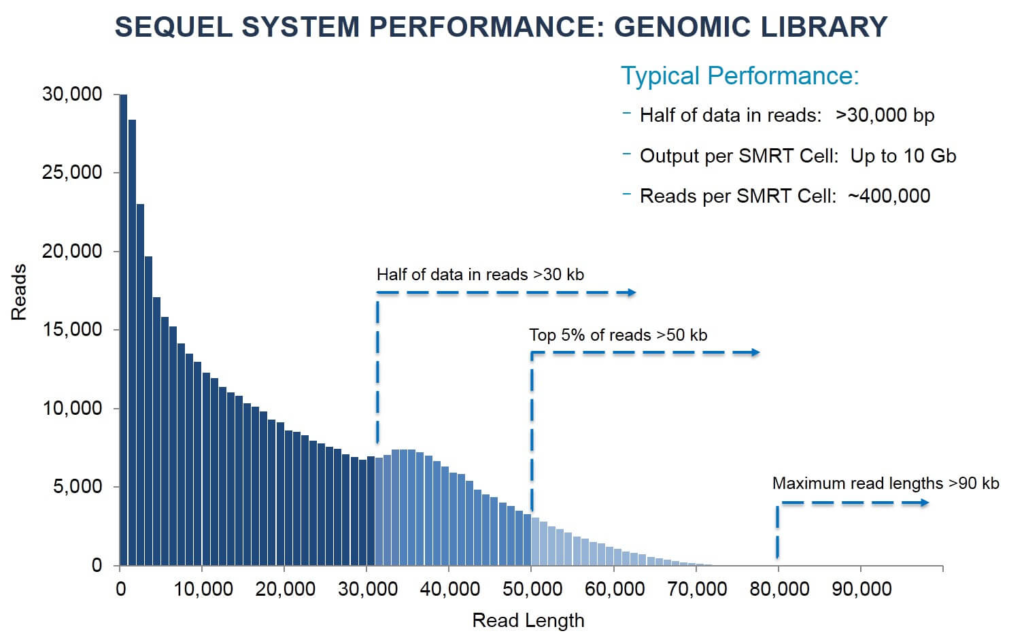
PacBio in numbers
(updated September 2017)
- RSII flowcell has 150,000 ZMWs, Sequel flowcell has 1,000,000 ZMWs;
- Typical RSII flowcell yield in ~1G base pairs of sequenced nucleotides; Sequel 5 - 8 Gbp.
- Optimal sequencing run yields in half or reads longer than 20k and some reads longer than 60k
Next
Learn about Oxford Nanopore Technologies: MinION .
Find out what Biological material we will be working with.
Go to page with tips, how to do a proper File organization .
If you know everything above, jump to Checkpoint .
Go back to Table of content .