Serratus Explorer - ababaian/serratus GitHub Wiki
Overview
Serratus Explorer (http://serratus.io/explorer) is the graphical front-end for accessing and exploring the data generated by the Serratus project. For programmatic access it is recommended you use the Tantalus R package interface. The explorer is designed to learn how the data is organized, and quickly validate findings based on systematic analysis.
There are two data-interfaces: nt contains nucleotide alignments for 3.8 million sequencing libraries aligned against a pangenome of Coronavirus sequences and RefSeq vertebrate viruses; rdrp contains protein alignments for 5.7 million libraries aligned against all known RNA dependent RNA Polymerases. This tutorial outlines nt usage as the example.
See also:
- SRA queries: SRA libraries included in Serratus searches (nt/rdrp)
- summary data format: Raw data formats
- Tutorial 1: How to find find novel viruses with Serratus.io
Navigation

Data can be explored by three main "keys";
1 - Family Level
Returns all "pangenome" entries belonging to nominal viral family across all SRA queries.
This search can be sub-set to return matches of varying nucleotide identity to reference sequences and based on score function. In brief, the score function is proportional to how many 'bins' of the pan-genome contain aligned reads.

Click "View Matches" and the SRA Runs containing a pangenome match to your query will be displayed 20 at a time. For "Identity = 80-95" and "Score = 80-100", there are 80 pages (~1600 matching SRA runs).
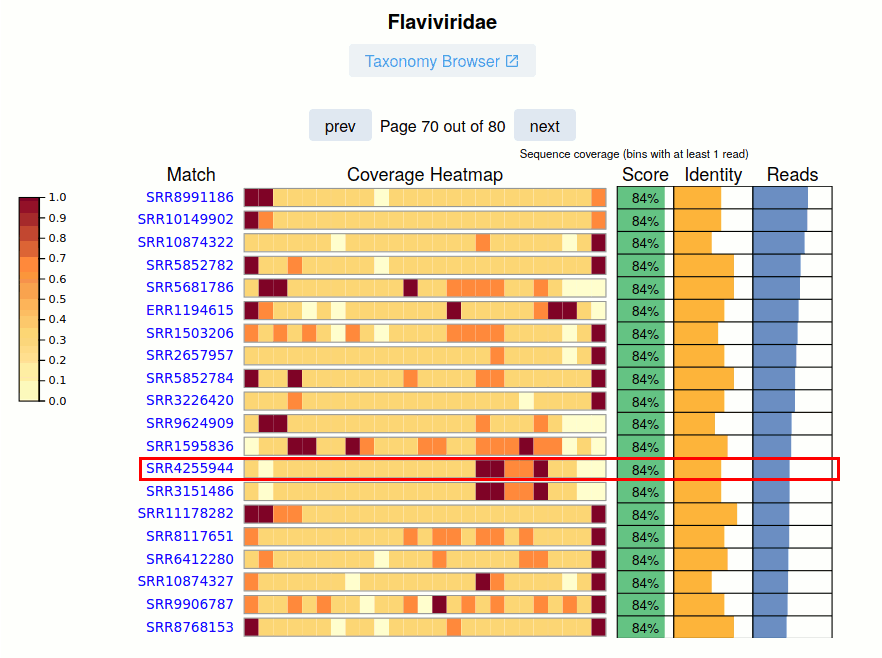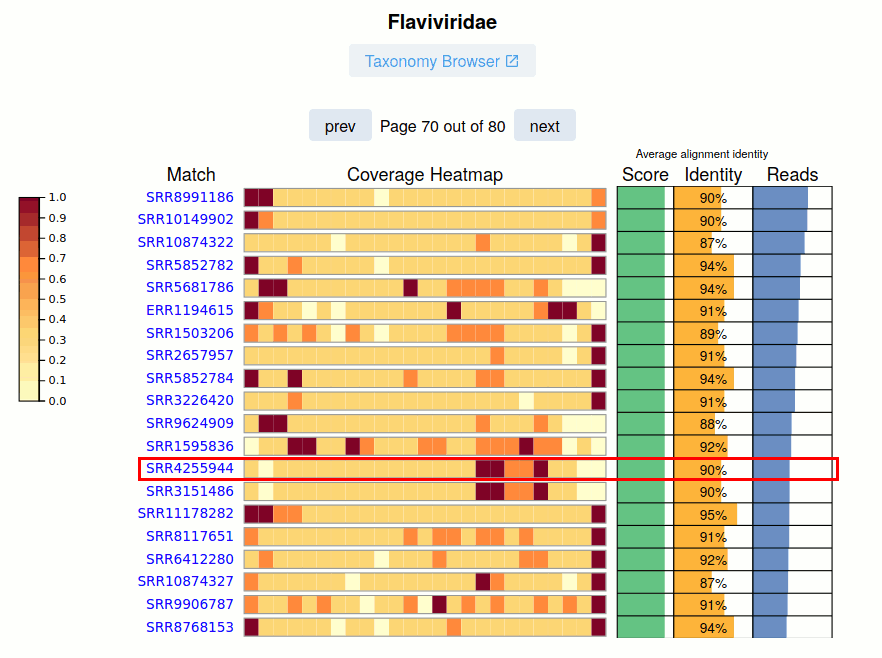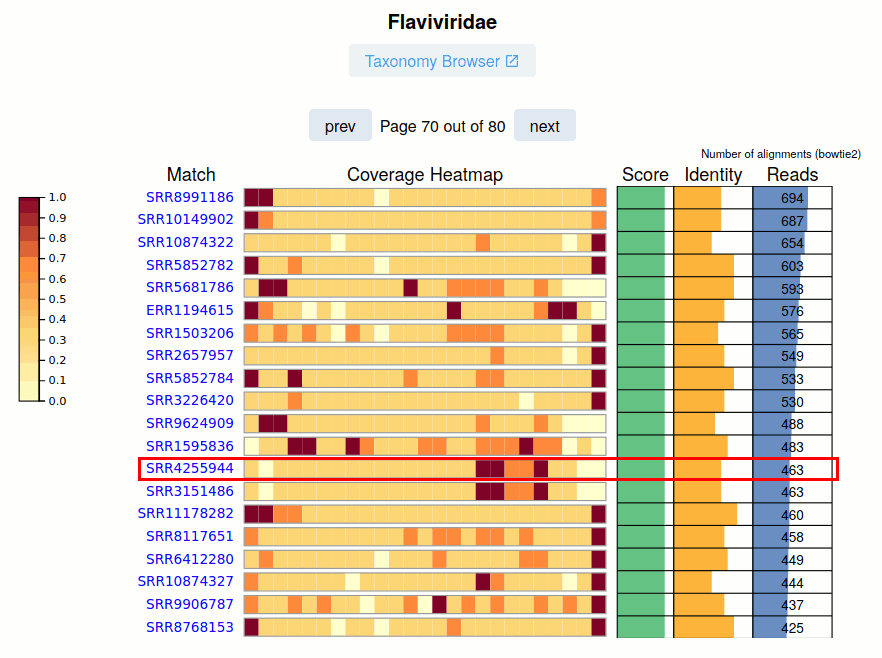
The SRA Run accession and link to the viral report is given.
The Coverage Heatmap is shown for the nominal viral pangenome. Currently this is represented as a coarse 'relative read distribution'.
The Pangnome Score, Average Read Identity, and Number of Aligned Reads for each SRA Run is provided on the right. Navigating these pages, especially at lower-identity ranges is a great way to uncover sequence biodiveristy.
See Also: Case Study Kenya Virome - SRR4255944
2 - GenBank Level
Return all SRA Runs containing reads mapped to a specific viral sequence included in the cov3ma pangenome.\
When you're seeking high-confidence matches (Score 80-100) this is a good way to identify relevant libraries.
A caveat with lower-confidence matches is that reads from another virus with sequence similarity can be 'mismapped' to the target sequence. For example, searching for NC_005148.1 Porcine Circovirus 2, returns SRA run "SRR9695950" with 118 reads at 88.8% identity. The Circoviridae pangenome for this library contains 50,839 mapped reads, primarily mapping to NC_001792.2 Porcine Circovirus 1. This type of "leaky alignment" is at times difficult to distinguish from novel viruses.

See Also: Sources of Error - Leaky Alignments
3 - SRA Run Accession Level
Return the complete viral summary report for a single SRA Run.
Both Family and GenBank searches ultimately link to individual SRA Run viral reports. This provides the richest data to infer on-target / off-target matches and an interface to view alignments directly for rapid 'validation'.
These are visualizations of the summary data format

For analysis see: Kenya Virome - SRR4255944
See also: Interpreting Error