4.4 Scoring - Valdes-Tresanco-MS/AMDock-win GitHub Wiki
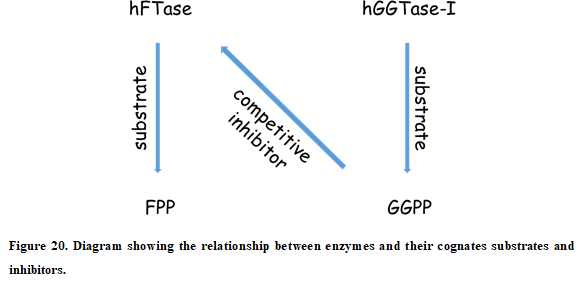
In this tutorial, we will study the effect of point mutations over ligand recognition by using the SCORING module of AMDock. We have selected as a study case the human farnesyl- transferase (hFTase), the same protein we used in the first tutorial. However, we will focus now on substrate (farnesyl diphosphate, FPP) recognition. The hFTase is a prenyl-transferase. It transfers a 15-carbon isoprenoid (donated by FPP) to the cysteine of a C- terminal motif of substrate proteins. Prenyl-transferases are generally highly selective for their cognate isoprenoid diphosphate substrates. Thus, hFTase binds FPP while the human geranylgeranyl-transferase type I (hGGTase-I, another prenyl-transferase) binds geranylgeranyl diphosphate (GGPP). The 20-carbon GGPP binds to hFTase with nanomolar affinity (but as a competitive inhibitor), while the 15-carbon FPP is a poor substrate for hGGTase-I.
Positioning GGPP-CaaX (GGPP + CaaX motif in proteins, where “a” is any amino acid) in the corresponding region of hFTase, leads to a steric clash with W102 and Y365 residues, suggesting that one or both of these residues may be key determinants of isoprenoid specificity. In elegant work, Terry et al.,1 re-engineered hFTase through site-directed mutagenesis to create an hFTase able to recognize GGPP substrate and geranylgeranylate its native CaaX substrates.

In this tutorial, we will try to predict the binding properties of both variants (the wild-type and the re-engineered) with AMDock SCORING module.
Remember, the SCORING module will not generate a new complex from those of the ligand and receptor coordinates. Essentially, this module calculates the binding energy between two molecules belonging to a complex by using the scoring function of the selected docking engine. So, the ligands and receptors coordinates we will use as input were extracted from those of the complexes X-ray structures (PDB IDs: 2H6F and 2H6G respectively).
Let’s begin with the wild-type first!
- Open AMDock program
- Select Autodock4Zn
- Set the Project Name (default Docking_Project)
- Set the Location for Project and push the button "Create Project"
- Check that “Scoring” checkbox is selected
- Choose the receptor file (Installation_path/Doc/Tutorials/III_Scoring/hFTase.pdb)
- Choose the ligand file (Installation_path/Doc/Tutorials/III_Scoring /FPP_CVLS.pdb)
- Press “Prepare Input” button
- As we said above, this module will not generate a new complex; hence, it is not necessary to define a search space. Press “Run” button then.
The predicted binding constant for the farnesylated CaaX peptide is 28nM. Let’s continue with the mutant…
- Open AMDock program
- Select Autodock4Zn
- Set the Project Name (default Docking_Project)
- Set the Location for Project and push the button “Create Project”
- Check that “Scoring” checkbox is selected
- Choose the receptor file (Installation_path/Doc/Tutorials/III_Scoring /hFTase_W102T.pdb)
- Choose the ligand file (Installation_path/Doc/Tutorials/III_Scoring /GGPP_CVLS.pdb)
- Press “Prepare Input” button
- Press “Run” button.
Here, the predicted binding constant for the geranylgeranylated CaaX peptide is 9.9nM which is, as expected, very similar to that of farnesylated CaaX peptide (28nM).