Link Particles to Tracks - UU-cellbiology/DoM_Utrecht GitHub Wiki
Creates tracks from detected particles/positions by linking them in consecutive frames. Requires Results table with detections to be opened.
Make notice, this option does not perform true nearest neighbor search. It is more simple and it was developed for quick linking of sparse datasets where small number of molecules per frame is present. Please use other plugins, like TrackMate or MTrackJ for complex and crowded cases. I plan to incorporate import/export of DoM_Utrecht to those plugins in the future.
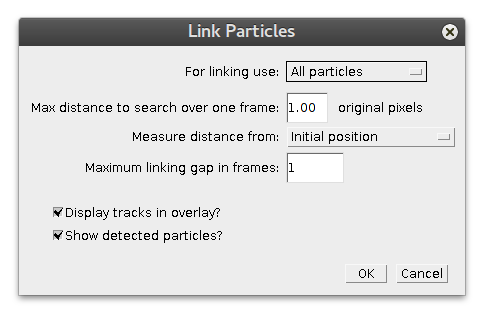
For linking use
defines which particles would be assembled into tracks, all or only true positives.
Max distance to search over one frame
Maximum displacement particle can have in track between consecutive frames. In case linking gap is allowed (so particle can "disappear" for a number of frames, see below), it is still the same distance over the missing time gap. Notice that this parameter is provided in pixels of original image.
Measure distance from
defines, what position in the current frame is used as a reference point for the search of particle in the next frame. If it is equal to Initial position, search for the next position would be carried out in vicinity of first particle's appearance. It can be handy for analysis of bleaching data, for example or if you want to highlight immobile particles.
For moving particles use Next detected position value, where next found position becomes new reference search position.
Maximum linking gap in frames
allows particle to "disappear" (or overcome missed detection) for specified number of frames. Note that next position will still be searched withing distance, defined by parameter above.
Display tracks in overlay?
adds randomly color-coded visualization of assembled tracks to the overlay of image. Requires original image stack to be open.
Show detected particles?
adds to overlay circles around detections used in linking. Requires original image stack to be open. It can be handy to quickly show detections without calling for Detect Molecules again.
So how does linking really works?
It sorts Results table by frame number and then starts linking process for the first particle in the table. All found "next positions" are removed from the list. If there is no particle in the vicinity at some point, new track is started from the next particle in the table. So there is no guarantee that found positions are "nearest neighbors", but if particles are sparse, it works quite well.