Detect Molecules - UU-cellbiology/DoM_Utrecht GitHub Wiki
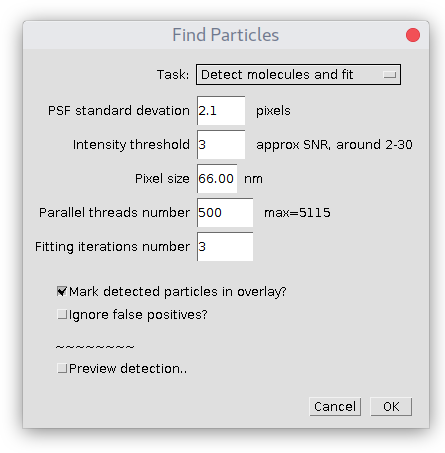
This is main analysis module detecting and fitting of 2D Gaussian shaped spots on image. It has following parameters:
Detection will be performed on whole image, unless some ROI is chosen in ImageJ. In this case only particles inside this ROI would be reported in Results table.
Spots that are close to the borders of an image (less than 3xSD of PSF) would be ignored.
Plugin uses Levenberg–Marquardt algorithm to fit 2D Gaussian with offset, minimizing the sum of least squares.
Task
Specifies what kind of job plugin should perform:
- Detect molecules (no fitting) task will just detect particles on image, but it will not perform fitting of 2D Gaussian. So that Results table will not contain any information about fitting (SD, localization error, etc) only X,Y coordinates of particles and their position within stack. X,Y coordinated are calculated from centroids, so the accuracy is quite good.
- Detect molecules and fit is default option. It will first detect molecules, perform Gaussian fitting and provide results.
- Fit detected molecules assumes there is already Results table present with detections in a proper format. This option is handy if detection is performed in some other plugin (for example, ComDet) and only fitting is required.
PSF standard deviation
The estimated standard deviation (width) of Gaussian in X and Y of Point Spread Function (PSF) of your microscope. Note, that this value must be provided in pixels of the original image. It does not have to be precise, it is used just as an initial fitting parameter and to optimize detection. If you don't know its value, you can read "How to quickly estimate SD of PSF in ImageJ" page to figure it out.
Intensity threshold
Allows the filtering of particles based on an estimated Signal-to-Noise ratio (i.e. remove dim particles). For better feeling use "Preview detection.." checkbox.
Pixel size
The physical dimension of one pixel in nanometers. Very important parameter, since all measurements are calculated using this scale. Check the magnification/camera pixel size of your setup or use the imaging of microscopic rulers to get this value.
Number of parallel threads
In short, this parameter defines how much memory plugin will occupy for its analysis. Specifically, if there is a stack of images, at every moment of time it will load to RAM and simultaneously process no more than this number of images from the stack. Use small numbers if you have little memory available (50-100 for 512Mb) and large if there is a lot of RAM (1000 for 1-10 GB). Easy to figure out its value empirically. But here is a real example of hard numbers, as illustration. Assume you have a stack of N=10000 images that are 16-bit and have width W=200 and height H=300 pixels. Total size of this image = 10000x200x300x16 bits or 10000x200x300x2 bytes, i.e. ~1.2Gb. If the number of threads is equal to 100 then the plugin will occupy 100x200x300x2 ~ 12 Mb of memory. Actually, it must be multiplied by a factor of 3 (since it reserves more for analysis), so it would be 36 Mb of RAM. Based on these calculations, using all available RAM, the maximum number of threads is estimated. A good initial value is about half of it.
Number of fitting iterations
Defines the maximum number of iterations performed during the fitting of 2D Gaussian to the shape of the individual molecule. From our experience, most of the fittings converge in about 6 iterations. If the quality of a fit is good enough, the plugin will stop earlier and report final numbers in Results table. Specifically, this parameter is the number of iterations in Levenberg–Marquardt algorithm mentioned above.
Mark detected particles in overlay
Adds round circles on top of detected particles to the overlay of analyzed image. Circle's color is green for "true positives" and red for "false positives". For big image stacks can be quite consuming for the memory, reserving some extra for displaying of overlay.
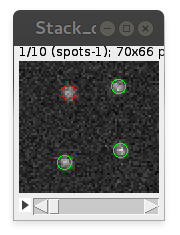
Ignore false positives
With this option checked, all "false positives" particles are ignored and not added to Results table.
Preview detection
Allows interactive visualization of detection on-the-spot. You can change PSF standard deviation and Intensity threshold parameters to find values for optimal detection. It support "detection inside ROI", if there is one selected before the start of the plugin.
How does detection works?
In short, detection/fitting scheme is presented on this picture:
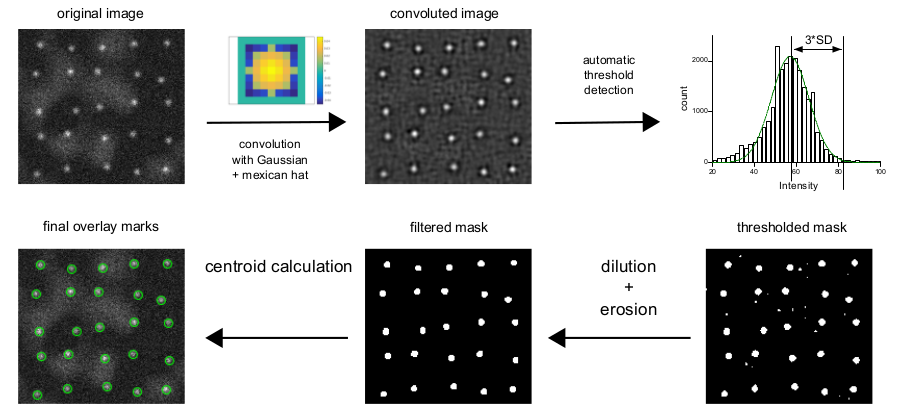
First, the image is convoluted with the Gaussian Mexican hat filter with the size provided by the user (PSF standard deviation, see above). Gaussian filter enhances spots of specified size, while the addition of "mexican hat" lobes removes the inhomogeneous background.
On the second stage, the plugin builds a histogram of the convoluted image and fits it with Gaussian to find a threshold. Gaussian fit estimates noise and average background value, i.e. majority of pixels at the image. From this, the threshold value of 3*SD (from fitting) is calculated.
Convoluted image is thresholded, some speckles are removed with dilation/erosion operations and so leftover spots are used to calculate centroid x,y on the original image. These values are used as initial parameters for 2D Gaussian fitting on the original image.