How to quickly estimate SD of PSF in ImageJ - UU-cellbiology/DoM_Utrecht GitHub Wiki
In ImageJ open image containing sparsely distributed single molecules acquired on your microscopy setup, so you can see individual emitters.
Make sure that you image does not have any scale, i.e. its width and height at the window title bar is displayed in pixels. If it has some physical units (um or nm), remove scale by Analyze - > Set Scale... -> Click to Remove Scale -> OK.
Using Straight Line Selection ROI tool draw a line across the spot of single emitter, approximately trying to cross pixel with maximum intensity:

Proceed to Analyze -> Plot Profile (or just press Ctrl+K)
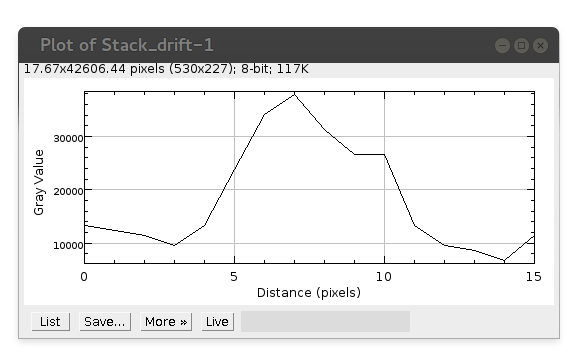
In this window press List button displaying intensity profile values:
Select all values (Ctrl+A) in the list and copy them to clipboard (Ctrl+C). Proceed to Analyze -> Tools -> Curve Fitting... and copy your values to this window, replacing existing one. Choose Gaussian in the dropdown list of available fitting functions:
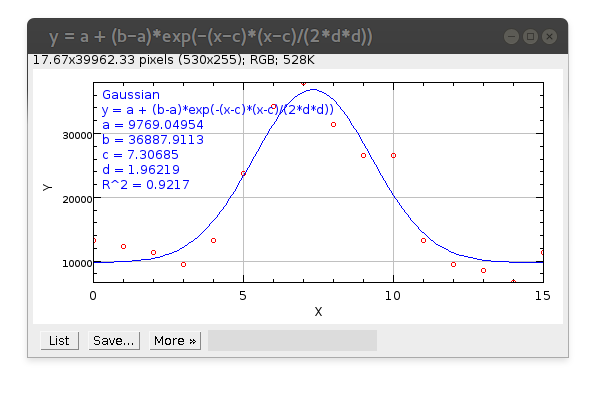
And press Fit button. New window should appear displaying good fitting of Gaussian curve (blue) to your data points (red circles):
If the fitting did not converge or looks bad, try repeating above steps using shorter or longer Line ROI.
The estimated SD of PSF parameter that is needed is d (in this specific example d=1.96219). You can use its value for Detection settings.
Ok, what about a bit more precise estimation using DoM plugin?
Yes, it is possible. Just analyze some image/stack with a lot of single molecules using Detect Molecules and go to Results table. Right click on any title of column and press "Distribution...". Choose Parameter: SD_X_nm, click OK and from displayed distribution pick average or mode value. Since it is in nanometers, to use it in DoM you would need to convert it back to pixels using pixel size of your microscopy setup.