Phase Contrast Cell Analysis Tool (Trainable WEKA Segmentation) - MontpellierRessourcesImagerie/imagej_macros_and_scripts GitHub Wiki
The tool allows to segment cells in non fluorescent microscopy images using the trainable WEKA segmentation. It allows to run a preprocessing that crops and converts images, to apply a classifier created with the Trainable Weka Segmentation plugin to a folder containing images and to open the images in a folder as a stack in the "Trainable Weka Segmentation plugin" to create a classifier.
You can find some example images here.
You can find an example classifier here 1_echantillon_quantif.model. You need to save the classifier into a folder models in your FIJI base folder.
Getting started
To install the tool, drag the link MRI - Phase Contrast Cell Analysis Toolto the FIJI launcher window, save it under macros/toolsets in the FIJI installation.
Select the "MRI - Phase Contrast Cell Analysis Tool" toolset from the >> button of the FIJI launcher.

- The first button (the one with the image) opens this help page
- The p-button runs the pre-processing (crop and convert the image type)
- The s-button runs the segmentation, applying a classifier from trainable the weka-segmentation
- The c-button opens the trainable weka plugin in order to create a classifier using your training images
Preprocessing
The pre-processing allows to crop the images in a folder and to convert them to 8-bit. Press the p-button to run the pre-processing. A dialog asking for the folder containing the images will be opened. The result images will be written to a subfolder "cropped" of the input folder.
Options
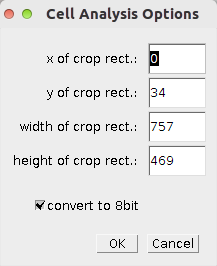
- x, y, width and height define the rectangle to which the image will be cropped
- select convert to 8bit if you want to convert the images to 8-bit.
Segmentation
Press the s-button to run the segmentation. A dialog that allows to select the folder containing the input images will be opened. Select the pre-processed images. A selection will be saved on each input image.
Options
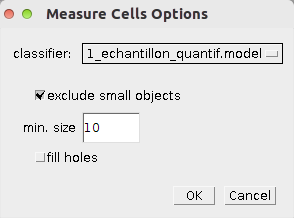
- Select the classifier you want to use from the list. The classifiers are taken from the folder models in the FIJI base folder
- exclude small objects - if selected objects smaller than a minimum size will be excluded from the result
- min. size - if exclude small objects is selected objects smaller than min. size are excluded from the result
- fill holes - if selected holes in detected objects are filled.
Create Classifier
The c-button allows to open the weka-plugin to create a new classifier. A dialog will be opened. Select the images that you want to use for the training of the classifier. They will be opened in the weka-plugin as a stack. When finished, save the classifier in the folder models under the FIJI base folder.
Results
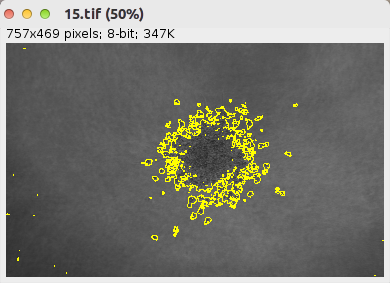

Publications citing the tool
-
Le Borgne-Rochet, M., Angevin, L., Bazellières, E., Ordas, L., Comunale, F., Denisov, E.V., Tashireva, L.A., Perelmuter, V.M., Bièche, I., Vacher, S., et al. (2019). [P-cadherin-induced decorin secretion is required for collagen fiber alignment and directional collective cell migration](https://jcs.biologists.org/content/132/21/jcs233189. J Cell Sci 132, jcs233189).
-
Planchon, D., Rios Morris, E., Genest, M., Comunale, F., Vacher, S., Bièche, I., Denisov, E.V., Tashireva, L.A., Perelmuter, V.M., Linder, S., et al. (2018). MT1-MMP targeting to endolysosomes is mediated by upregulation of flotillins. Journal of Cell Science 131, jcs218925.