restriction digestion - MetabolicEngineeringGroupCBMA/MetabolicEngineeringGroupCBMA.github.io GitHub Wiki
When working with enzymes it is important to plan you experiment before taking out the enzymes from the freezer. When out, keep them in a stratacooler.

Return the enzymes to the -20 freezer as soon as you are done.
In order to perform one restriction digestion, add the components in the table below together in the indicated order.
| Component | Volume (µL) |
|---|---|
| Sterile ddH2O water | 16.5 |
| Restriction Enzyme Buffer(10x) | 2 |
| Plasmid DNA ~1-2 µg | 1.0 |
| Restriction enzyme (10 U/µL) | 0.5 |
Final volume should be 20 µL. Mix gently by pipetting, close the tube and centrifuge for a second to collect the liquid at the bottom of the tube.
Incubate at the enzyme's optimum temperature (usually 37°C) for 1 hour.
Note: Overnight digestions are usually unnecessary.
Gel electrophoresis of the DNA is necessary in order to find out if the digestion was successful.
It is important to run an undigested control on the gel. Remember to always load an equal amount of DNA in each well of the agarose gel.
The best way of making this control is to make an identical mix like the one above, but with water instead of the restriction enzyme. This control should be incubated at the same temperature as the restriction digestion.
If you suspect problems, add a control DNA that should be digested.
If you need to digest more than one or two different DNA preparation, it is better to prepare a restriction digestion master mix in analogy of a PCR mastermix.
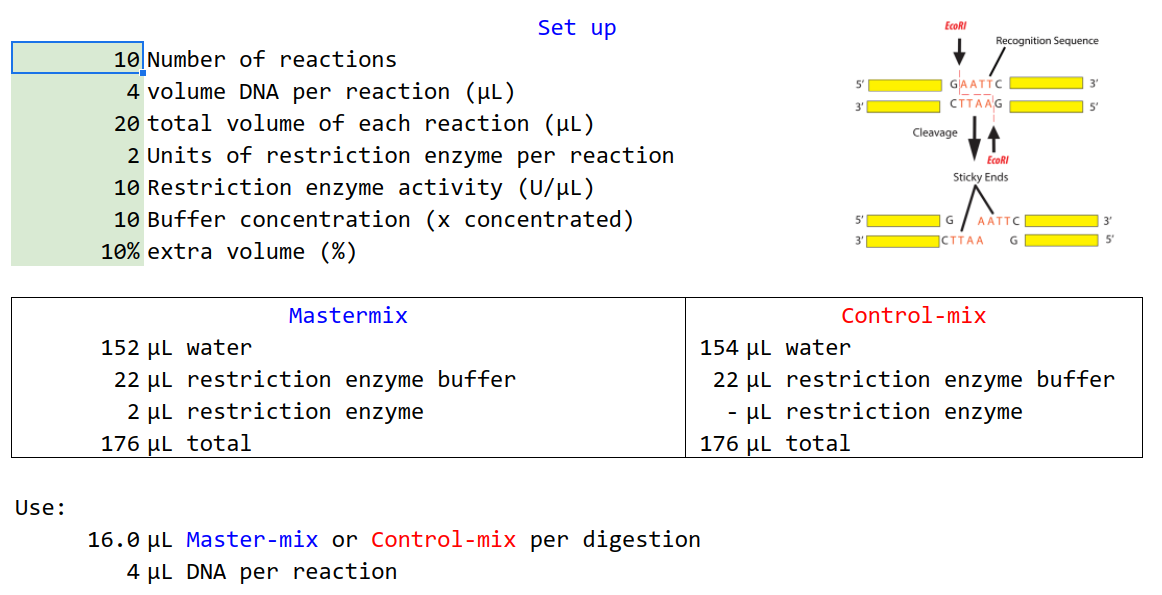
Imagine if you would like to set up ten digestions of 20 µL with 4 µL of DNA in each.
In this example, your restriction enzyme has 10 U/µL and you would like to have 2 U per reaction. The restriction buffer is x10 concentrated and you would like to make ten percent more than you need to make up for pipetting errors.
The table below lists the initial relevant variables.
| Value | Variable |
|---|---|
| 10 | Number of reactions |
| 4 | volume DNA per reaction (µL) |
| 20 | total volume of each reaction (µL) |
| 2 | Units of restriction enzyme per reaction |
| 10 | Restriction enzyme activity (U/µL) |
| 10 | Buffer concentration (x concentrated) |
| 10% | extra volume (%) |
Prepare the Mastermix like so:
| Volume (µL) | Component |
|---|---|
| 152 | water |
| 22 | buffer |
| 2.2 | restriction enzyme |
| 176 | total |
Use the mastermix as described in the table below. It is easier to add the mastermix in all the tubes first, since the DNA has a low volume.
| Volume (µL) | Component |
|---|---|
| 16.0 | Mastermix per digestion |
| 4 | DNA per reaction |
The volumes of this mastermix was calculated using this google spreadsheet (shown below). The green fields are variables you can change.
You can modify directly according to your needs or download it in excel format.
| Component | Volume (µL) |
|---|---|
| Unpurified PCR product | 25 |
| Restriction enzyme | 0.5 |
No restriction buffer is added to the digestion of PCR products. This works for enzymes that show activity in PCR buffers
This protocol i useful if you need to digest a PCR product for some analytical purpose. However, even after thermal cycling is complete, the thermostable DNA polymerase retains some activity, which could fill in the sticky ends generated during a subsequent restriction digestion. These blunt-ended products can result in reduced cloning efficiency.
Digesting a DNA substrate with two restriction enzymes simultaneously (double digestion) can save a lot of time. Some restriction enzymes come with a universal buffer such as the FastDigest from Thermo Scientific. If you are using enzymes with specific buffer buffers, there are online tools such as NEBcloner that will guide you to select conditions.
Under non-standard reaction conditions, some restriction enzymes are capable of cleaving sequences which are similar, but not identical, to their defined recognition sequence. This effect is called star activity. One common cause of this is too high glycerol content in the reaction mix. The final reaction mix should not contain more than 5% v/v glycerol.
Restriction enzymes are stored in 50% glycerol, therefore the amount of enzyme added should not exceed 10% of the total reaction volume. Remember to account for alkaline phosphatase is present. More on star activity here.
Quote from the net: "One thing I also noticed when I use the Thermo Fisher Double Digest Tool: Nearly every enzyme has around 50 percent activity in BamHI Buffer. I very often use the BamHI Buffer for double digests that do not work for the conventional color buffers. Maybe FD buffer is similar to BamHI Buffer. Try out BamHI Buffer, if your double digests do not work."
We use thermosensitive alkaline phosphatases, such as Shrimp alkaline phosphatase (SAP), FastAP or Arctic phosphatase.
Using an alkaline phosphatase to remove phosphates from the ends of linear DNA and may improve ligation and gap repair since it inhibits self ligation of the empty vector.
These enzymes are active in restriction buffers, so you do not need to add any specific buffer. It also works well in PCR buffer. The enzymes can be inactivated by 5-15 min at 65 - 75 °C according to the manufacturers specification. It is important to perform this inactivation prior to ligation as the enzyme will otherwise remove phosphates from the insert as well, which inhibits ligation. See this BiteSize Bio blog post for more information on the use of phosphatases in cloning.
Find restriction sites in your sequence using Restriction Analyzer or RestrictionMapper. Learn more about Restriction Enzymes, this site has very good general information from PDB.
Learn how to identify Supercoils, Nicks and Circles in Plasmid Preps from this BiteSize Bioblog post.