User Function Definitions and the Processing Stream - BUNPC/Homer3 GitHub Wiki
This page offers a detailed explanation of Homer3's processing stream and the format of a user function.
Homer3 can import properly formatted functions for use in a processing stream, making it easy for MATLAB users to customize their analysis and share their methods with other users of the software. This guide first describes how the user function registry works, how the processing stream works, and then explains how to format a function for use with Homer3.
Unfortunately, only users with MATLAB who use Homer3 source code can create and edit user functions.
The Processing Stream
It is important to understand how the processing stream is executed in order to write custom user functions.
In Homer3, analysis is carried out by a series of user functions that are each a part of a processing stream. When these functions are defined to have arguments and return values which share the same names, these variables are manipulated over the course of the processing stream’s execution, allowing multiple functions to generate the desired output in series.
Thus, the names and formats of these input and output variables, which are defined by the Homer3 documentation and standards such as the SNIRF file specification, are critically important. A table of the most frequently used processing stream inputs, outputs, variables, and their formats is available here.
A simple example: The following run level processing stream converts raw intensity to optical density, and then to concentration data. Then it calculates block averages of each signal following the stim marks:
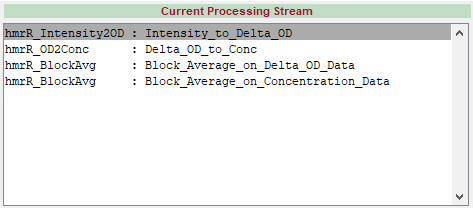
User Functions
User functions are the building blocks of a processing stream. Each function should carry out a modular bit of analysis on a type of fNIRS data or its derivatives. User functions exist for filtering signals, pruning data or stims based on quality metrics, and carrying out sophisticated statistical analyses.
The Function Registry
User functions must be imported to the function registry before they can be used in a processing stream. The function registry is generated when Homer3 is run for the first time. It can be regenerated by selecting Reload Registry from the File menu of the Edit Processing Stream window. An individual script can be added to the function registry by selecting Import User Function from the File menu of the Edit Processing Stream window. User function scripts are loaded from the Homer3\FuncRegistry\UserFunctions folder. Each function should be defined in its own script with the same name, otherwise it will not be recognized when the registry is reloaded.
Usage Options
Often, we would like to be able to use the same function with various sets of inputs and outputs: statistical analysis and filtering is not usually specific to any single type of data. However, recall that the literal names of the variables passed into and returned from user functions define the type of data the function operates on. Homer3 user functions resolves this by allowing users to define multiple usage options for a function. Usage options are defined in the user function help string; see below.
See the Usage Options for hmrR_BlockAvg:
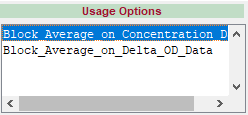
There is a version of the function for use with concentration data and another for use with optical density, each with their own set of inputs and outputs. We can see the usage of each function in our processing stream by clicking on its name in the Current Processing Stream Window. At the most basic level, the processing stream is simply the execution of each function’s selected usage option, in order from top to bottom:
dod = hmrR_Intensity2OD(data)
dc = hmrR_OD2Conc(dod, probe, ppf)
[dcAvg, dcAvgStd, nTrials, dcSum2] = hmrR_BlockAvg(dc, stim, trange)
[dodAvg, dodAvgStd, nTrials, dodSum2] = hmrR_BlockAvg(dod, stim, trange)
Note that the OD2Conc function does not destroy its input, dod, which is still available for the block average even after being “converted” to dc.
The processing stream also manages the arguments to the functions and their return values. Each of these can be classified as an input, an output, or a parameter.
Inputs and Outputs
Variables which Homer3 makes available before the processing stream has been run, such as stimulus marks, imported SNIRF data, and lists of channels and timepoints to be excluded or included in analysis, are called processing stream inputs:

When the processing stream finishes running, dod, dc, and any other variables returned by the processing stream will be saved to disk within groupResults.mat. These are called processing stream outputs:

When users define their own inputs and outputs, they show up under “misc” by default.
Parameters
Not all arguments passed to the functions in the example above come from the processing stream inputs or the output of previous functions; namely, ppf in hmrR_OD2Conc and trange in hmrR_BlockAvg. These arguments are called parameters, which means they are defined by the user via the Edit Options dialog.
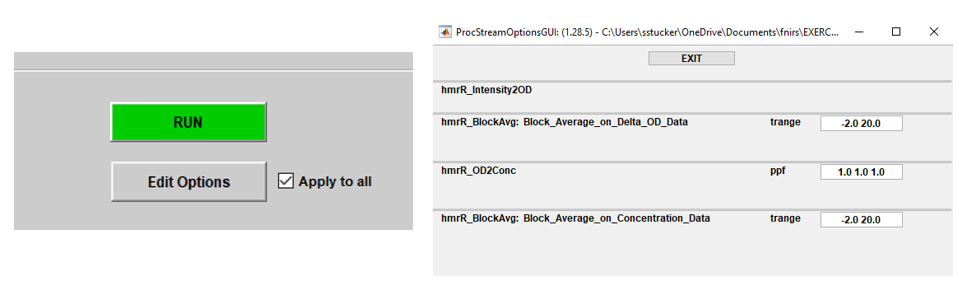
Hovering over a parameter’s name in this GUI will display a tooltip which offers details about the parameter and the values it can take. Homer3 knows which arguments are parameters and what their default values should be because it is defined in the user function help string.
Defining a User Function
User functions are more than just MATLAB functions: user function definitions include meta information such as documentation, the names of parameters and their default values and usage options. All this information is written in the user function help string, which is parsed by Homer3 when the function is imported into the registry.
User functions must be the first function in their own .m file, and must be located in the Homer3\FuncRegistry\UserFunctions folder, or a subfolder of this directory.
The Help String
The help string is broken into 7 parts. Each part is required.
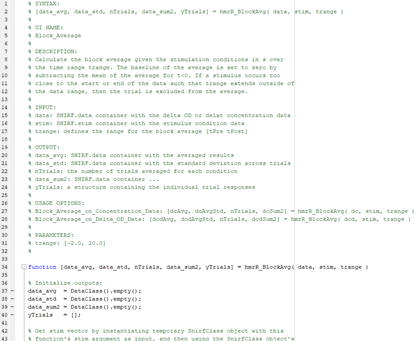
The SYNTAX section of the help string should mirror the actual function which is defined in the .m file.
% SYNTAX:
% [data_avg, data_std, nTrials, data_sum2, yTrials] = hmrR_BlockAvg( data, stim, trange )
The string in the UI NAME section will appear in places in the GUI where a short token that is not specific to the selected usage option is appropriate for display.
% UI NAME:
% Block_Average
The string in the DESCRIPTION section will appear as a tooltip when hovering over the function name and will appear in Edit Processing Stream GUI when the function is selected.
% DESCRIPTION:
% Calculate the block average given the stimulation conditions in s over
% the time range trange. The baseline of the average is set to zero by
% subtracting the mean of the average for t<0. If a stimulus occurs too
% close to the start or end of the data such that trange extends outside of
% the data range, then the trial is excluded from the average.
All arguments to the function must be listed in the INPUT section, line by line. Function inputs must be made available by Homer3 before the processing stream is run or be an output of a previously run function or be defined as a parameter in the PARAMETERS section below. A colon should precede a short description of the input’s intended format. If the argument is a parameter, the description should detail the values the parameter can take and what behaviors these values correspond to. Note that these names are the names that will appear in various tool tips and GUI panels, but do not need to be the literal variable name if there are multiple usage options.
% INPUT:
% data: SNIRF.data container with the delta OD or delat concentration data
% stim: SNIRF.stim container with the stimulus condition data
% trange: defines the range for the block average [tPre tPost]
Outputs of the function must be listed in the OUTPUT section. A colon should precede a short description of the output’s format. Note that these names are the names that will appear in various tool tips and GUI panels, but do not need to be the literal variable name if there are multiple usage options.
% OUTPUT:
% data_avg: SNIRF.data container with the averaged results
% data_std: SNIRF.data container with the standard deviation across trials
% nTrials: the number of trials averaged for each condition
% data_sum2: SNIRF.data container ...
% yTrials: a structure containing the individual trial responses
The USAGE OPTIONS section lists the literal calls that can be made to the function. (see above for an explanation of Usage Options) The usage options defined here must use the correct variable names or the function will not be called correctly.
% USAGE OPTIONS:
% Block_Average_on_Concentration_Data: [dcAvg, dcAvgStd, nTrials, dcSum2] = hmrR_BlockAvg( dc, stim, trange )
% Block_Average_on_Delta_OD_Data: [dodAvg, dodAvgStd, nTrials, dodSum2] = hmrR_BlockAvg( dod, stim, trange )
Function arguments which are parameters must be listed in the PARAMETERS section line by line. A colon should precede the default value of the parameter. The way the default value is listed defines the format of the parameter and thus the way it is displayed, loaded, and saved. The number of decimal points in the provided default value determine the number of decimal points that will be displayed, loaded, and saved. i.e. a parameter with a default value of 1 that should be able to take the value of 0.99 should be given a default value of 1.00. If the parameter is a matrix of multiple numbers, the numbers should be surrounded in brackets. Matrix parameters must have a fixed maximum length.
% PARAMETERS
% trange: [-2.0, 20.0]