Typical processing streams - BUNPC/Homer3 GitHub Wiki
Homer3 enables you to create your own processing stream files in the Process Stream GUI. This document provides typical processing streams that are used to analyze fNIRS datasets in the Homer3 toolbox.
The basic processing streams typically include four functions:
a) hmrR_Intensity2OD: converts raw data to optical density;
b) hmrR_BandpassFilt: performs a band-pass filter on optical density data;
c) hmrR_OD2Conc: converts the filtered optical density to hemoglobin concentration;
d) hmrR_BlockAvg: and finally calculates the average hemodynamic response function (HRF).

You can also estimate the HRF using general linear model (GLM) analysis by incorporating the GLM function in the basic stream. The following processing stream provides typical processing and parameters for short-separation regression using the GLM approach, typically including four functions:
a) hmrR_Intensity2OD: converts raw data to optical density;
b) hmrR_BandpassFilt: performs a band-pass filter on optical density data;
c) hmrR_OD2Conc: converts the filtered optical density to hemoglobin concentration;
d) hmrR_GLM: and finally estimates the HRF using GLM analysis.
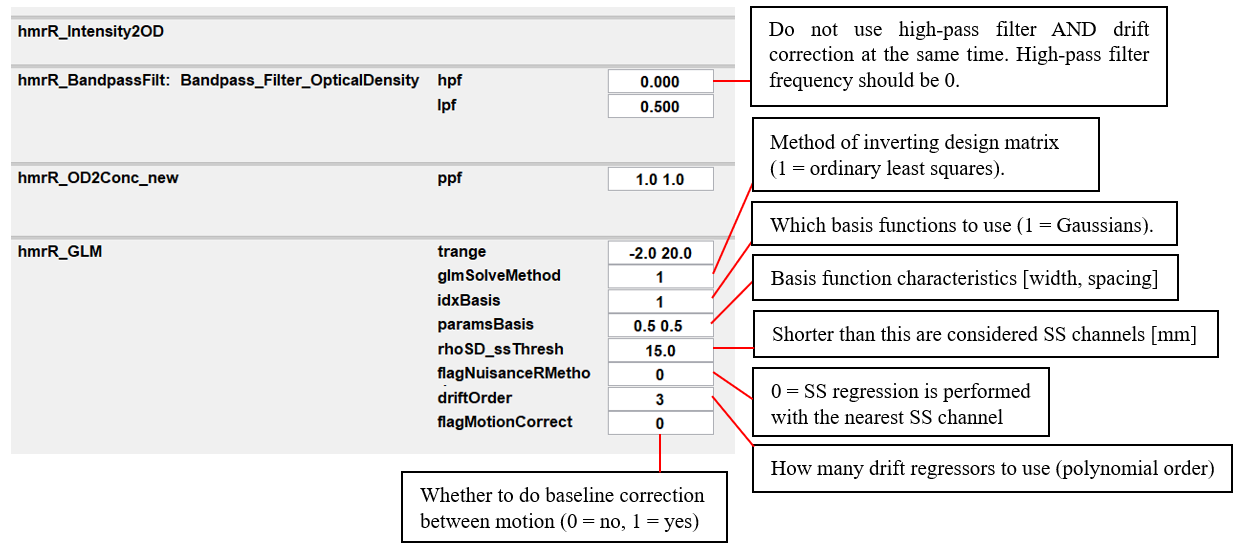
Here, these parameter settings build a design matrix with Gaussian basis functions, 3 drift regressors AND with a short separation regressor based on the nearest SS channel.
fNIRS allows measurements in relatively unrestrained environments and is prone to motion artifacts (MAs). Homer3 enables you to identify MAs and discard MA-contaminated trials from your analysis.
The following is a typical processing stream including the functions of pruning channels, identifying and rejecting MAs.
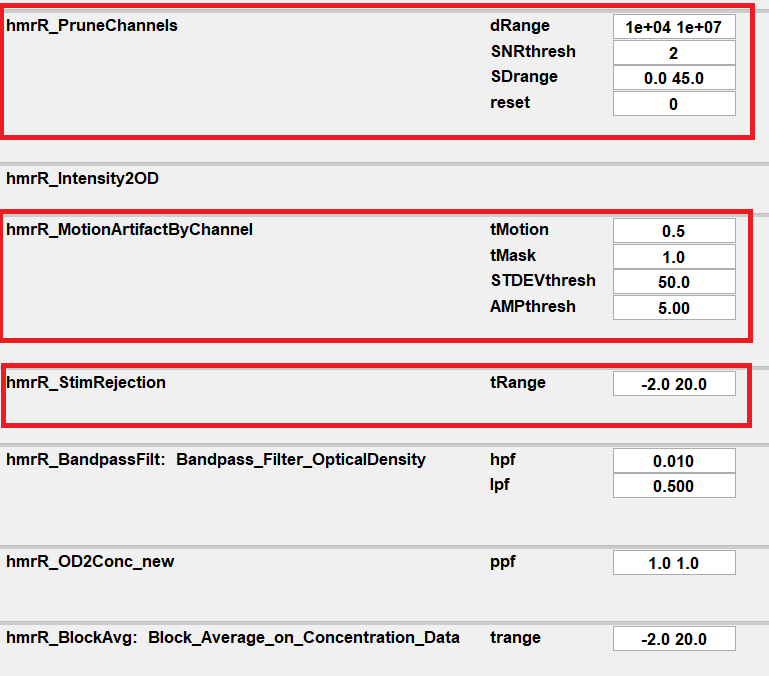
hmrR_PruneChannels is used to prune channels from the measurement list if their signal is too weak, too strong, or their standard deviation is too great. Whether the data meets these conditions depends on the parameters of this function: dRange, SNRthresh, SDrange, and reset.
- dRange – If mean(data) < dRange(1) or > dRange(2) then it is excluded as an active channel. Default: dRange(1)=1e+04, dRange(1)=1e+07.
- SNRthresh – If mean(data)/std(data) < SNRthresh then it is excluded as an active channel. Default: 2.
- SDrange – prune channels with a source-detector separation < SDrange(1) or > SDrange(2). Default: SDRange(1)=0, SDRange(2)=45.
- reset – reset previously pruned channels (automatic and manual).
hmrR_MotionArtifactByChannel performs channel-wise motion artifacts identification in an input data matrix d. If any active data channel exhibits a signal change greater than STDEVthresh or AMPthresh, then a segment of data around that time point is marked as a motion artifact.
- tMotion– Check for signal change indicative of a motion artifact over time range tMotion. Units of seconds.
- tMask– Mark data over +/- tMask seconds around the identified motion artifact as a motion artifact. Units of seconds.
- STDEVthresh– If the signal d for any given active channel changes by more that STDEVthresh × stdev(d) over the time interval tMotion, then this time point is marked as a motion artifact. The standard deviation (stdev) is determined for each channel independently.
- AMPthresh– If the signal d for any given active channel changes by more that AMPthresh over the time interval tMotion, then this time point is marked as a motion artifact.
hmrR_StimRejection excludes from HRF calculation the stimuli that fall within the time points of segments identified as motion artifacts. Its parameter tRange is an array of two numbers [t1 t2] specifying how many seconds surrounding motion artifacts. Typical values are t1=-2 and t2 equal to the stimulus duration. The stimulus is the time-0 point. In this example, tRange = [-2 20] means if there is a motion artifact within 2 seconds before and 20 seconds after a stimulus, the block corresponding to this stimulus is excluded from the block-averaging to compute the HRF.
Motion artifacts will be marked with pink patches, and excluded stimuli will appear as doted lines in the Homer3 GUI, as shown in the below figure.
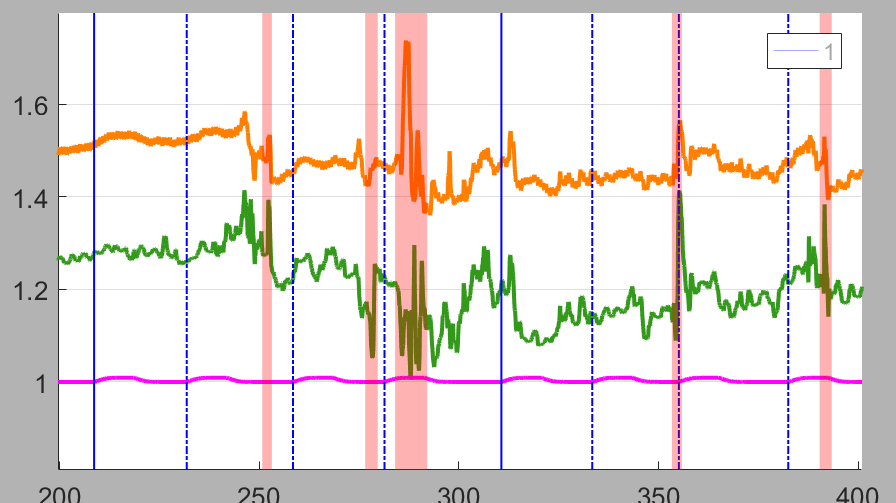
Note: When computing the HRF by block-averaging (hmrBlockAvg), the stimulus blocks containing motion artifact will only be excluded if explicitly instructed with the StimRejection function. When computing the HRF by GLM analysis (hmrGLM), the uncorrected MA segments will be excluded from the analysis.
If you have a limited number of trials, motion artifact correction would be a better option than StimRejection. There are multiple advanced functions you can incorporate in the processing stream to perform motion artifact correction, such as MotionCorrectSpline, MotionCorrectPCA, MotionCorrectPCArecurse, MotionCorrectWavelet, and MotionCorrectCbsi.
Here gives the processing stream containing all MA correction options and their typical parameters.

It should be noted that there are advantages and limitations for each method of MA correction. For example, Spline motion correction is a simple, fast approach that can correct signal offsets and MA segments, but requires a reliable technique to identify segments and sometimes results in overfitting. PCA filter is very efficient when motion is the dominant source of variance (such as for infants) but tends to remove too much of the activation signal in adults. Wavelet performs better at maintaining frequency content, in particular keeps physiology, but this method does not correct signal offsets and works slower than other approaches.
The appropriate method mainly depends on the type of motion artifacts.