Hadley Wickham R Packages - statnet/computing GitHub Wiki
[Hadley Wickham's R Packages Book] (http://r-pkgs.had.co.nz)
- Names
- Letters and numbers only recommended, mixing upper and lowercase not ideal
- Abbreviations, adding extra R, and words referencing the function or problem
- Creating
- Can use RStudio File|New Project|New Directory|R Package|Create Project
devtools::create("path/to/package/pkgname")
- Components
- R/ directory
-
DESCRIPTIONfile -
NAMESPACEfile
- Created
.Rprojtext file- Use Project Options within the projects menu
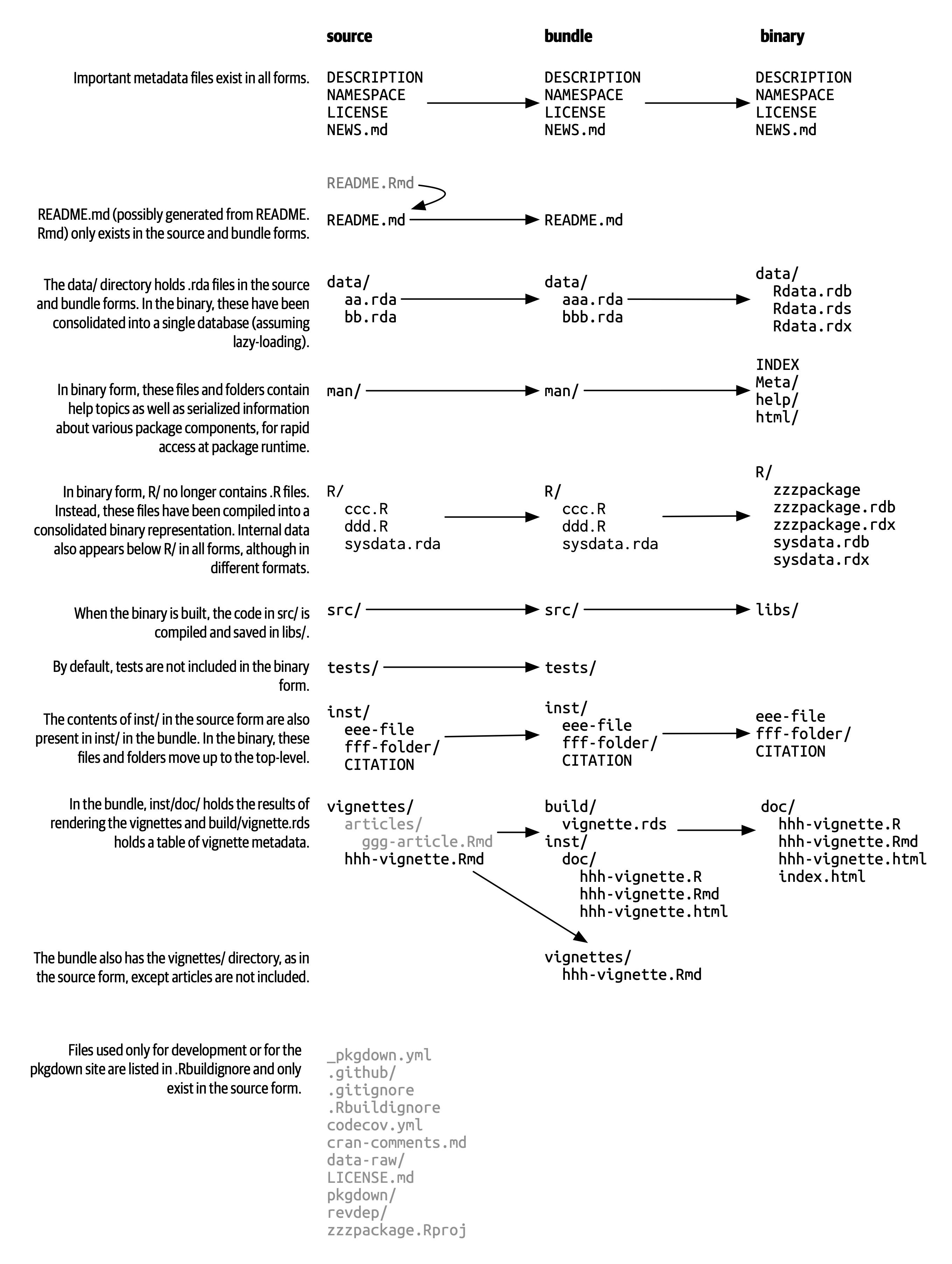
- Package Types
- Source: development version
- Just the 3 main components
- Bundled: compressed into a single file
.tar.gz-
.Rbuildignoreuseful for CRAN packages, prevents files in source from appearing in bundle
-
- Binary: for distribution to users without package development tools
- Installed: binary package decompressed into a package library
- In memory: a package loaded into memory
- Source: development version
- Library
- Used to load packages
-
Organizing functions
- Can use
devtools::load_all()to source all files in R/ - Don't put all functions into one file, and don't put each function in its own file
- Can jump to definition of a function
- Click the function name and press F2
- Press Ctrl and start typing the name
- Can use
-
Code style
- Good coding can save time and lives
- formatR package works well to fix problems, lintr package works well to warn
- Object names
- Underscore for separation, lowercase
- Spacing
- Space around arithmetic operators and in function calls ( = )
- : :: ::: are exceptions to this
- Space before left parentheses, except in a function
- Can extra space to improve alignment
- Space around arithmetic operators and in function calls ( = )
- Curly braces
- Opening brace should not be on its own line, but closing brace should unless followed by
else
- Opening brace should not be on its own line, but closing brace should unless followed by
- Line length: 80 characters per line
- Indent: use 2 spaces
- If a function runs over multiple lines, then indent to where the definition starts
- Use <- to assign, rather than an equals sign
- Comment using #, but can also use - and = with # to make code more readable
-
Scripts and packages
-
Don't run code at the top level of a package
- One example is to create a function solely for output (graphs), and a separate one for data prep
- Scripts: code is run when loaded
- Package: code is run when built --> should only create objects
- Other people will use your package
- Don't use
source() - Don't use
library()orrequire()
- DESCRIPTION file can be used to describe necessary packages for your code
-
Don't run code at the top level of a package
-
Side effects for packages
- Good for when package talks to an external system
-
.onLoad()(Most common) and.onAttach() - Custom options
- Defining s4 classes, methods, and generics
- DESCRIPTION file
- Started with `devtools::create("mypackage")
- Package, Title, Version, Authors@R, Description, Depends, License, and LazyData fields
- Dependencies
- Add using
devtools::use_package("dplyr", "Suggests")ordevtools::use_package("dplyr") -
Imports(must be present)- Adding a dependency ensures installation, but not attachment to the package
- Refer to external functions with
package::function()
-
Suggests(can be used but not required)- Not automatically installed
requireNamespace("pkg", quietly = TRUE)
- Can specify versions of packages needed (usually stick to a minimum version)
dplyr (>= 0.3.0.1)
- Add using
- Other specialized dependencies
-
Dependsbetter used in namespaces -
LinkingTopackages rely on C or C++ -
Enhancesrarely used, but means that you provide methods for classes in another package
-
-
Title: One line plain-text description (65 characters) -
Description: One paragraph- Recommended to also include a README.md file
-
Authors@R: author and maintainer -
License: Important for releasing -
Version: Integers (major.minor.patch.dev) (.dev is dropped when released) -
Collate: Order of sourcing -
LazyData: Increase ease of data access
- Started with `devtools::create("mypackage")
-
Reference documentation (
.Rdfiles) accessed by?orhelp()- Use roxygen2 package to turn formatted comments into
.Rdfiles- Can also manage
NAMESPACEandCollatefield inDESCRIPTION
- Can also manage
- Use roxygen2 package to turn formatted comments into
-
Workflow
- Add roxygen comments
- Run
devtools:document()(or press Ctrl/Cmd+Shift+D in RStudio) to convert - Preview with
?and repeat
-
Formatting for roxygen comments
- Start with
#'and are called blocks - Blocks are broken into tags, which look like
@tagName details - Text before first tag is introduction
- First sentence is a one-line title
- Second paragraph is brief description
- Third and other paragraphs go into details
- Can add arbitrary
@section -
@seealsocan point to other resources either on the web,\url{http://www.r-project.org}, in your package\code{\link{functioname}}, or another package\code{\link[packagename]{functioname}}. -
@familyfor linking to related functions -
@aliasesadds more names for the topic that can be used with? -
@keywordscan be useful for developers, particularly@keywords internal
- Start with
#' Add together two numbers.
#' @param x A number.
#' @param y A number.
#' @return The sum of \code{x} and \code{y}.
#' @examples
#' add(1, 1)
#' add(10, 1)
#' \dontrun{
#' add("a", "b")
add <- function(x, y) {
x + y
}
- Output after running
devtools:document()
-
Alternative Workflow
- Add roxygen comments
- Click Build and Reload in the build pane or press Ctrl/Cmd+Shift+B
- Preview with
?and repeat
-
Documenting functions
-
@param-
@param name descriptiondescribes inputs or parameters- Should start with a capital letter and end with a full stop
- Can document multiple arguments with commas, using
@param x,y Numeric vectors
-
-
@examples- Provides executable R code, that doesn't necessarily need be in the documentation (can specify a pathname)
- Good to include code that doesn't run
\dontrun{}
-
@return-
@return descriptiondescribes the output
-
-
-
Documenting packages
- Accessed with
package?foo - Can supplement vignettes
- No object for a package, so need document
NULLand label it with@docType packageand@name <package-name>
- Accessed with
-
Documenting classes, generics, methods
- S3 generics are regular functions and S3 classes have no formal definition, and methods are optional
- S4 generics are regular functions,
- S4 classes should have a roxygen block before
setClass(), use@slotto document slots in the same way that@paramwas used for functions - S4 methods must be documented in either the class, generic, or in its own file
- Ordering for S4
- Code needs to run in a certain order (rather than default alphabetical)
-
@includein roxygen2 says that one file must be loaded before another
- Reference classes use docstring
-
Special characters includes
\to escape -
Inherit parameter descriptions from another function using
@inheritParams source_functionor@inheritParams function -
Documenting multiple functions in the same file
- Can be confusing, best used when functions have similar arguments or complementary effects (
open()andclose()) -
@describeInis most common -
@rdnamecan overwrite default file names or add documentation to existing function@name
- Can be confusing, best used when functions have similar arguments or complementary effects (
#' Foo bar generic
#' @param x Object to foo.
foobar <- function(x) UseMethod("foobar")
#' @describeIn foobar Difference between the mean and the median
foobar.numeric <- function(x) abs(mean(x) - median(x))
#' @describeIn foobar First and last values pasted together in a string.
foobar.character <- function(x) paste0(x[1], "-", x[length(x)])
- Text formatting in
.Rd-
Character
-
\emph{italics}: italics. -
\strong{bold}: bold. -
\code{r_function_call(with = "arguments")}:r_function_call(with = "arguments")(format inline code) -
\preformatted{}: format text as-is, can be used for multi-line code
-
-
Links
-
\url{http://rstudio.com}: a url. -
\href{http://rstudio.com}{Rstudio}:, a url with custom link text. -
\email{hadley@@rstudio.com}(note the doubled @): an email address.
-
-
List
- Ordered
-
#' \enumerate{
#' \item First item
#' \item Second item
* Unordered
#' \itemize{
#' \item First item
#' \item Second item
- Definition
#' \describe{
#' \item{One} First item
#' \item{Two} Second item
- Mathematics can use LaTex
\eqn{a + b}for inline or `\dqn{a + b} for display (block) - Tables
-
\tabular{}- Column alignment argument (1=left, r=right, c=centre)
- Table contents with columns separated by
\taband rows by\cr
-
First argument
-
tabular <- function(df, ...) {
stopifnot(is.data.frame(df))
align <- function(x) if (is.numeric(x)) "r" else "l"
col_align <- vapply(df, align, character(1))
cols <- lapply(df, format, ...)
contents <- do.call("paste",
c(cols, list(sep = " \\tab ", collapse = "\\cr\n ")))
paste("\\tabular{", paste(col_align, collapse = ""), "}{\n ",
contents, "\n}\n", sep = "")
}
cat(tabular(mtcars[1:5, 1:5]))`
- Second argument
## \tabular{rrrrr}{
## 21.0 \tab 6 \tab 160 \tab 110 \tab 3.90\cr
## 21.0 \tab 6 \tab 160 \tab 110 \tab 3.90\cr
## 22.8 \tab 4 \tab 108 \tab 93 \tab 3.85\cr
## 21.4 \tab 6 \tab 258 \tab 110 \tab 3.08\cr
## 18.7 \tab 8 \tab 360 \tab 175 \tab 3.15
## }
- Long-form documentation that can describe and show solutions to a problem
-
browseVignettes()Source file, readable HTML or PDF, and a file of R code - knitr uses rmarkdown
-
-
devtools:: use_vignette("my-vignette")- Creates directory, adds necessary dependencies, and drafts a vignette
- Made of 3 components
- Metadata block
- Title, author, date, output, vignette
- Markdown for formatting
- --- for horizontal rules
-
- for unordered lists, 1. for ordered, and : for definition lists
- _ or * for italics, __ or ** for bold
- [link text](destination) or <http://www.google.com> for links
- ` for code, ``` for block code
- \ for escaping
- Knitr
- Similar to a fenced code block, runs the code, captures the output, and translates it
- Metadata block
\```{r}
# Add two numbers together
add<-function(a, b) a + b
add(10, 20)
- Can specify options to control rednering
*
eval = FALSEprevents evaluation *echo = FALSEturns off printing of code input *results = "hide"turns off printing of code output *error = TRUEcaptures and displays errors inlin *collapse = TRUEandcomment = "#>"to display output *results = "asis"treats R code output as markdown *fig.show = "hold"holds figures until end of block *fig.width = 5and `fig.height = 5' for height and width
- Automated tests are important
-
devtools:: use_testthat()- Creates a directory (
tests/testthat), adds testthat to theSuggestsfield, and creates a file in which tests are run - Test using Ctrl/Cmd + Shift + T or `devtools::test()
- Dots are passed tests, numbers are failed tests
- Creates a directory (
-
Expectations grouped into tests which are organized in files
- Expectations: right value and right class? error messages produced? Binary
-
expect_equal: equality within a numerical tolerance -
expect_identicalfor exact equivalence or comparing environments -
expect_match()matches character vectors against regular expression-
expect_output()for printed output,expect_message()for messages,expect_warning()for warnings,expect_error()for errors -
expect_is()checks inheritance from a certain class
-
-
- Tests: groups multiple expectations, created with
test_that() - Files: groups multiple related tests, created with
context()
- Expectations: right value and right class? error messages produced? Binary
- Skip
- Prints an s in the output rather than throwing an error
check_api <- function() {
if (not_working()) {
skip("API not available")
}
}
test_that("foo api returns bar when given baz", {
check_api()
...
})
library(stringr)
context("String length")
test_that("str_length is number of characters", {
expect_equal(str_length("a"), 1)
expect_equal(str_length("ab"), 2)
expect_equal(str_length("abc"), 3)
})
test_that("str_length of factor is length of level", {
expect_equal(str_length(factor("a")), 1)
expect_equal(str_length(factor("ab")), 2)
expect_equal(str_length(factor("abc")), 3)
})
test_that("str_length of missing is missing", {
expect_equal(str_length(NA), NA_integer_)
expect_equal(str_length(c(NA, 1)), c(NA, 1))
expect_equal(str_length("NA"), 2)
})
- Spaces for names - a context for looking up values of objects associated with names
-
::operator is one example to reference the right package -
Imports: defines how a function in one package finds a function in another
- Controls which external functions can be used by your package without having to use
:: - Recommended to note package name in
Imports:field inDESCRIPTIONand call functions usingpkg::fun() - Repeated use of functions - can import using
@import package - S3 generics follow same rules as functions, and methods are also available when generic is imported
- S4 classes:
@importClassesFrom package ClassA ClassB - S4 generics:
@importMethodsFrom package GenericA GenericB @import methods
- Controls which external functions can be used by your package without having to use
-
Exports: avoid conflict with other packages by specifying which functions are available outside of your package
-
@exportin the roxygen comment block will generate exports - Export functions for others to use, better to export too little than too much
- Can export S3 generics like functions, S3 classes by
@exportthe constructor function - S3 methods: differs by exported/internal generic in a required/suggested package (4 scenarios)
- S4 generics and methods:
@export - S4 classes and RC:
@exportif you want others to be able to extend, otherwise@exportthe constructor function
-
-
search()to look for all packages attached- **Loading **loads code and data, registers methods, and runs
.onLoad() - **Attaching ** puts the package in the search path
-
library()andrequire()load and attach the package
- **Loading **loads code and data, registers methods, and runs
- Use
library()in data analysis, never in packages (useDependsorImportsfields in theDESCRIPTIONfile)-
Dependsloads the package,Importsattaches it - Always list packages in
Dependsrather thanImportsunless there is a good reason, such as being used in conjunction with another package
-
- Use `requireNamespace(x, quietly = TRUE) in a package if a specific action is needed based on the presence/absence of a suggested package
- Recommended to generate
NAMESPACEwith roxygen2- Workflow
- Add roxygen comments to your .R files.
- Run
devtools::document()(or press Ctrl/Cmd + Shift + D in RStudio) to convert roxygen comments to.Rdfiles. - Look at NAMESPACE and run tests to check that the specification is correct.
- Workflow
- Can store binary data and make it available to user (
data/), store parsed data but not make it available (R/sysdata.rda), or store raw data (inst/extdata)-
data/-
.Rdatafile created bysave()with a single object -
devtools::use_data()ordevtools::use_data_raw()
-
-
- Alternatively, can include it in the source of your package
- Document using roxygen2
-
@formatgives an overview of the dataset after a title and description -
@sourceprovides details of where data was obtained from -
Never
@exporta data set
-
- Internal data
- Save in
R/sysdata.rda devtools::use_data( , internal = TRUE)
- Save in
- Raw data
- Include original files in
inst/extdata - Use
system.file()to refer
- Include original files in
- C or C++ are faster than R, Rcpp package
-
devtools::use_rcpp()- Creates a
src/directory to hold.cppfiles - Add
Rcppto theLinkingToandImportsfields in theDESCRIPTION. - Set up a
.gitignorefile to make sure you don’t accidentally check in any compiled files (learn more about this in git). - Tell you the two roxygen tags you need to add to your package:
@useDynLib your-packageand@importFrom Rcpp sourceCpp
- Creates a
-
- Workflow
- Create a new C++ file from File, which will include a default template
-
#includesand// [[Rcpp::export]]most important
- Generate the necessary modifications to your NAMESPACE by documenting them with Ctrl/Cmd + Shift + D.
- Click Build & Reload in the build pane, or press Ctrl/Cmd + Shift + B.
- Run
timesTwo(10)from the console to check that it works.
- Documentation
- Each exported function gets a wrapper function
- Base function
.Call()to execute a function
timesTwo <- function(x) {
.Call('timesTwo', PACKAGE = 'mypackage', x)
}
- Can use roxygen2 to document this like R, but use
//for comments, rather than#
//' Multiply a number by two
//'
//' @param x A single integer.
//' @export
// [[Rcpp::export]]
int timesTwo(int x) {
return x * 2;
}
-
Exporting C++ code
- To make C++ callable,
// [[Rcpp::interfaces(r, cpp)]]
- To make C++ callable,
-
Importing C++ code
- In
DESCRIPTION, addLinkingTo: otherPackage - In the C++ file, add:
#include <otherPackage.h> - Use
otherPackage::foo()to access functions
- In
-
Best practices
- Print output using
Rcout << ... - In long-running loops, regularly run
Rcpp::checkUserInterrupt() - Use
.hextension for headers and include files - writing a
.onUnload()function that unloads the DLL to clean up:
- Print output using
.onUnload <- function (libpath) { library.dynam.unload("mypackage", libpath) }
* Use clang instead of gcc to compile your C++ code
#### C
* C works with older packages or if it is an existing C library
* Call C functions using `.C()` and `.Call()`
* Include two header files
#include <R.h> #include <Rinternals.h>
* `SEXP` is short for S expression
SEXP add_(SEXP x_, SEXP y_) { double x = asReal(x_); double y = asReal(y_);
double sum = x + y;
return ScalarReal(sum); }
#' @useDynLib mypackage add_ add <- function(x, y) .Call(add_, x, y)
* `.C()`
* First write a void C function
void add_(double* x, double* y, double* out) { out[0] = x[0] + y[0]; }
* Then create an R wrapper
#' @useDynLib mypackage add_ add <- function(x, y) { .C(add_, x, y, numeric(1))3 }
* Workflow
1. Modify the C code.
2. Build and reload the package with Ctrl/Cmd + Shift + B
3. Experiment at the console.
* Exporting C code
* **Function registration** to solve relocatable DLL issue
* `R_RegisterCCallable()` for export, `R_GetCCallable()` for import
* Export has a pointer to the DLL, function name, and a pointer to the function
```
#include "add.h"
#include <R_ext/Rdynload.h>
void R_init_mypackage(DllInfo *info) {
R_RegisterCCallable(info, "add_", (DL_FUNC) &add_)
}
```
* Import has function name and package name
```
#include <R_ext/Rdynload.h>
#include <R.h>
#include <Rinternals.h>
SEXP add_(SEXP x, SEXP y) {
static SEXP(fun*)(SEXP, SEXP) = NULL;
if (fun == NULL)
fun = (SEXP(*)(SEXP, SEXP)) R_GetCCallable("add", "mypackage");
return fun(x, y);
}
```
* Importing C code
* If it uses the system described above, all you need is `LinkingTo: otherPackage` in the `DESCRIPTION`, and `#include otherPackageAPI.h` in the C file.
* If it registers the functions, but doesn’t provide a header file, you’ll need to write the wrapper yourself. Use `Imports` and not `LinkingTo`. You also need to make sure the package is loaded. You can do this by importing any function with `@importFrom mypackage foo`, or by adding `requireNamespace("mypackage", quietly = TRUE)` to `.onLoad()`
* Best Practices
* Avoid calls to `assert()`, `abort()1 and `exit()`: Instead, use `error()` which is equivalent to calling `stop()` in R
* To print output use `Rprintf()`, not `printf()`
* In long-running loops, regularly call `R_CheckUserInterrupt()`
* Don’t use C’s random number generators (like `rand()` or `random()`), instead use the C API to R’s rngs: `unif_rand()`, `norm_rand()`, etc. Note the caveats in “Random number generation” - you must call `GetRNGstate()` before and `PutRNGstate()` after
* Use R macros `ISNAN(x)` and `R_FINITE(x)` to check for NaNs and infinite values
* Unload the DLL when the package is unloaded:
```
.onUnload <- function (libpath) {
library.dynam.unload("mypackage", libpath)
}
```
* Use `clang` instead of `gcc` to compile your C code
#### Debugging compiled code
* Open a shell and start R
R --debugger=lldb
R --debugger=gdb
* Start R with `process start` (lldb) or `run` (gdb)
* Now in an interactive debugger
* `help` for a list of all commands
* `thread backtrace/bt` for location, Navigate the callstack with `frame select <n>/frame <n>`, or `up` and `down`
* Evaluate the next expression with `thread step-over/next`, or step into it with `thread step-in/step`. Continue executing with `thread step-out/finish`
* Show all variables defined in the current frame with `frame variable/ info locals,` or print the value of a single variable with `frame variable <var>/p <var>`
* Makefiles
* Use `Makevars`
* `PKG_LIBS`: Linker flags
* `PKG_CFLAGS` & `PKG_CXXFLAGS` for C and C++ flags, set define directives with `-D`
* `PKG_CPPFLAGS`: Pre-processor flags, set directories with `-I`
## Installed files
* When a package is installed, everything in `inst/` is copied into the top-level package directory
* Don't use subdirectories with the same names as existing directories
* `inst/AUTHOR` and `inst/COPYRIGHT`: If the copyright and authorship of a package is particularly complex, you can use plain text files inst/COPYRIGHTS and inst/AUTHORS to provide more information
* `inst/CITATION`: how to cite the package, see package citation for details
* `inst/docs`: This is an older convention for vignettes, and should be avoided in modern packages
* `inst/extdata`: additional external data for examples and vignettes. See external data for more detail.
* `inst/java`, `inst/python etc` . See other languages
* `citation()` for base R or for packages
## Other components
* Rare directories
* `demo/` for package demos, `exec/` for executable scripts, `po/` for translations, `tools/` for auxiliary files
* Demos show how to combine multiple functions to solve a problem `demo()`
* **Vignettes are preferred over demos**
## Git and GitHub
* Git is a version control system for tracking code, GitHub is a website
install.packages("devtools") devtools::install_github("username/packagename")
* Can call a few Git commands from RStudio, but should use **shell**
* RStudio: Tools>Shell
* Commands rather than functions used in shell
* R = `f(x, y = 1)`, shell = `f x --y=1` or `f x -y1`
* `pwd`: print working directory
* `cd <name>`: change working directory
* `ls`: list files
1. Install Git
2. Tell Git your name and email
3. Create a GitHub account
4. Generate a SSH key and give public one to GitHub
* Create a local Git repository
* RStudio > Tools > Global Options > Git/SVN panel
* Run `git init` in a shell
* You'll see a git pane at the top right and a git dropdown menu
* Can now see what has changed, can `git status` to get overview or `git diff` to show differences
* Unit of work is called a **commit** and is a snapshot of code at one timepoint
* Unique identifier (SHA), changeset, commit message, a parent, and an author
* You **stage ** files (tell Git what changes should be included) and then commit those files (Click *Commit* or press Ctrl + Alt + m)
* Select files to be staged and write a commit message
* In a shell `git add` stages new and modified files, `git rm` stages deleted files, and `git commit -m <message>` commits
* Commit best practices
* Minimal and complete, concise and evocative, the why not the what
* Ignoring files
* Add files to `.gitignore` by right-clicking on file in Git pane and choosing *Ignore*
* Undo mistakes
* Right-click on file in Git pane and select *Revert*, or click *Discard chunk* in the diff window
* If not right away, need to look back by clicking *History* in the Git pane
* Navigate back in time to find the commit and write down the parent SHA
* `git show <SHA> <filename>
* Sync with GitHub
* Create a new repo with GitHub with the same name as your package
* Open a shell
```
git remote add origin [email protected]:hadley/r-pkgs.git
git push -u origin master
```
* Modify `DESCRIPTION` to add `URL` and `BugReports` fields that link to new GitHub site
* Save file and commit, then **Push** changes
* **Push** sends changes to GitHub, others will need to **pull*, which fetches changes and then merges them, which can cause conflict
* If a conflict exists:
git merge --abort
git config --global merge.conflictstyle diff3
git pull
* Don’t fix problems in man/*.Rd files. Instead, resolve any conflicts in the underlying roxygen comments and re-document the package.
* Merge conflicts in the NAMESPACE file will prevent you from re-loading or re-documenting a package. Resolve them enough so that the package can be loaded, then re-document to generate a clean and correct NAMESPACE.
* Issues tracker
* Exists on GitHub, can assign tasks, can be opened and closed
* Branches
* Default is `master`, `origin/master` if you sync with GitHub
* Can be useful when you want to make big changes or break away from main stream
* Create with `git checkout -b <branch-name>`
* Switch branches with `git checkout <branch-name>
* `git branch` for a list of all
* To sync with GitHub, run this: `git push --set-upstream origin <branch-name>`
* To merge a branch back and then delete it:
```
git checkout master
git merge <branch-name>
git branch -d <branch-name>
```
* Pull request
* To contribute to someone's code
* Create a branch, commit code, then push the branch to GitHub
* Submitting to another repo
* **Fork** the original repo to create a copy belonging to you
* **Clone** the forked repo to create a local copy
```
git clone [email protected]:<your-name>/<repo>.git
cd <repo>
```
* Keep forked and original repo in sync:
```
git remote add upstream [email protected]:<original-name>/<repo>.git
git fetch upstream
git merge upstream/master
git branch -u upstream/master
git checkout master
git pull
git checkout <my-branch>
git merge master
```
## Checking
* Foundation is `R CMD check` but *don't call directly*
1. `devtools::check()` or Cmd + Shift + E in RStudio
* Ensures up-to-date documentation, bundles before running, skips tests on CRAN
2. Fix problem and repeat
* Returns `ERROR`, `WARNING`, `NOTE`
* You check many things
* Metadata
* Packages: directory, source package, executable files, hidden files and directories, portable file names, sufficient permissions, package installation, package size, top level files, subdirectories, left-over files
* DESCRIPTION: meta-information, dependencies, CRAN feasbility
* NAMESPACE: existence, information, loading with stated dependencies
* Code: non-ASCII characters, syntax errors, dependencies, S3 generic/method consistency, replacement functions, possible problems, package and namespace loading and unloading
* Data: directory
* Documentation: `devtools:: check_doc()`, check Rd files, metadata, line widths, cross-references, missing documentation entries, code/documentation mismatches, `\usage`, contents, unstated dependencies, examples, manual
* Demos: index information
* Compiled code: foreign function calls, line endings, compiled code
* Tests: unstated dependencies, tests
* Vignettes: 'build' directory, installed files, vignettes, PDF file size, unstated dependencies, package vignettes, running R code, re-building of vignette outputs
* Check with Travis-CI
* Runs automated testing code every time you push to GitHub
* Create a Travis account
* `devtools::use_travis()`
* Enable Travis for the repo you want to test from your account
* Commit and push to GitHub
* `r_github_packages` and `r_binary_packages`, which give lists of packages to install
## Releasing a package
1. Pick a version number
* Increment patch for bug fixes without new features
* Increment minor for new features, changes in backwards compatibility
* Increment major for non-backwards compatibility
2. Run and document R CMD check
3. Check that you’re aligned with CRAN policies
4. Update README.md and NEWS.md
5. Submit the package to CRAN
6. Prepare for the next version by updating version numbers
7. Publicize the new version
* Backwards compatibility
* Incompatibility can be good to fix design mistakes, but changes should be gradual
* Don't immediately remove a function, start by deprecating `.Deprecated`
* Warn in the function using `warning()`
* Can add a helper function to automatically display a message, warning, or error
* Check for the correct version at run-time
if (packageVersion("ggplot2") < "1.0.0") { stop("ggplot2 >= 1.0.0 needed for this function.", call. = FALSE) }
if (packageVersion("foo") > "1.0.0") { foo::baz() } else { foo::bar() }
* Submission to CRAN
* `cran-comments.md` should be created and checked into git and listed in `.Rbuildignore`
* Create a package bundle with `devtools::build()` and upload to CRAn
* Test environments (Mac, Windows, Linux, Solaris) - at least two
* Install **R-devel** on your own machine
* Travis can run checks on linux
* Check results
* Reverse dependencies if a new version of an existing package `devtools::revdep_check()`
* CRAN policies
* Stable email address for maintainer
* Identify copyright holders in `DESCRIPTION`
* Package working across multiple platforms
* No external changes without user permission
* README.md and README.Rmd
* Why should I use, how should I use, how should I get it?
* `devtools::use_readme_rmd()`
* Must re-knit `README.Rmd` every time you modify it
* NEWS.MD
* Lists API changes in each release
* `devtools::use_build_ignore("NEWS.md")`
* Release
* `devtools::release()`: builds and re-runs `R CMD check` one more time and asks check questions