Heatmap and UMAP - simoncmo/pik3ca_proteomics GitHub Wiki
Tutorial to use heatmap/UMAP
- to start using the function, source all files using
## Source functions
walk(list.files('script/proteomics/src/', full.names = T), source)
PANCAN
# Step1: Make Plot object
pancan_obj = MakeDataObj(protein_mtx_human, meta = meta, top_var = 0.8) %>%
MakeCluster(k=5) %>%
MakeUmap()
Various heatmap
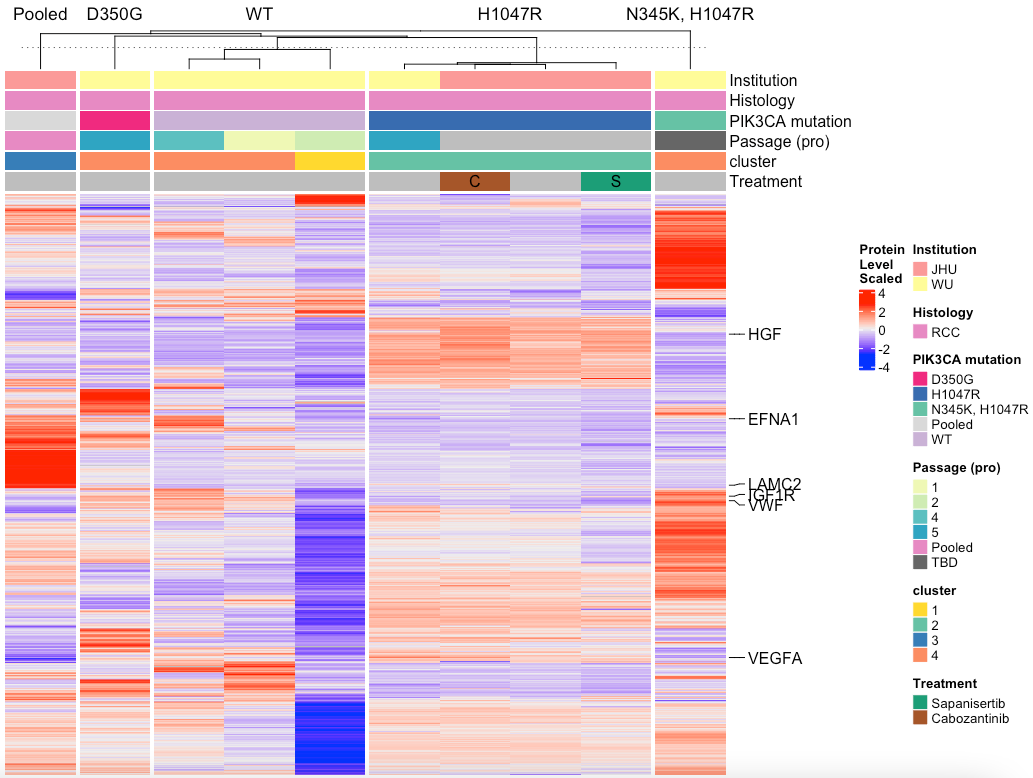
MakeTopProHeatmap(pancan_obj, palette_list = palette_list_pancan, split_column = F,
anno_pch_cols = c('Histology', "Treatment"),
anno_regular = c("Institution","PIK3CA mutation","Passage (pro)", 'cluster'))
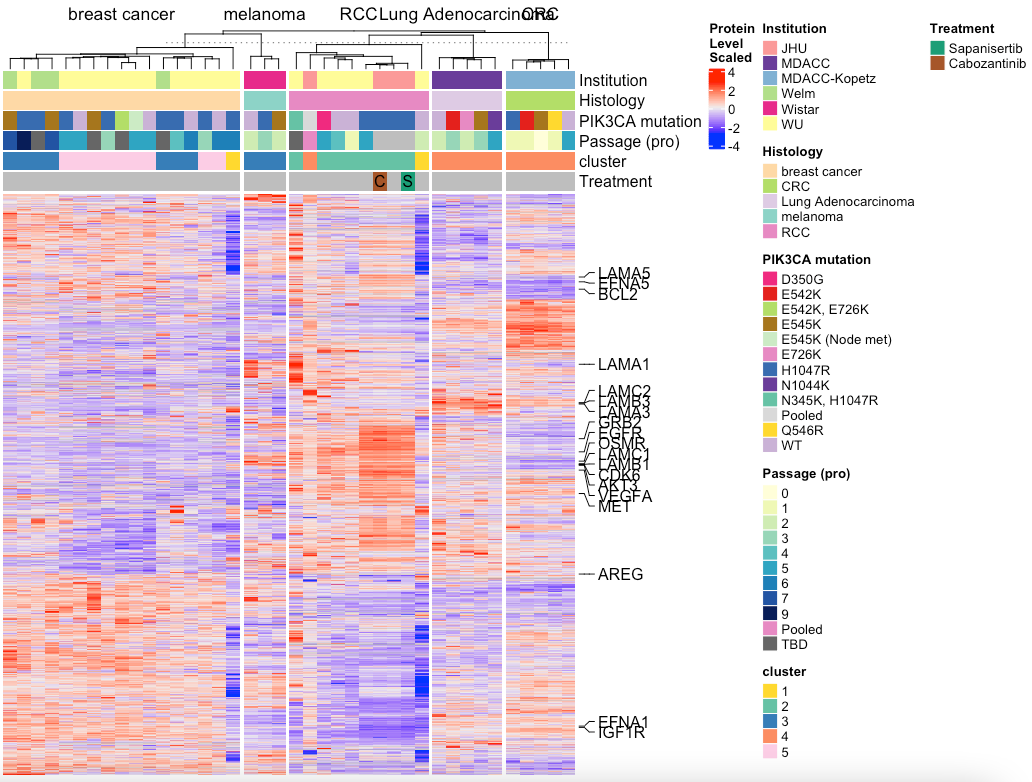
# Split by histology
MakeTopProHeatmap(pancan_obj, palette_list = palette_list_pancan, split_column_by = 'Histology')
# Split by cluster
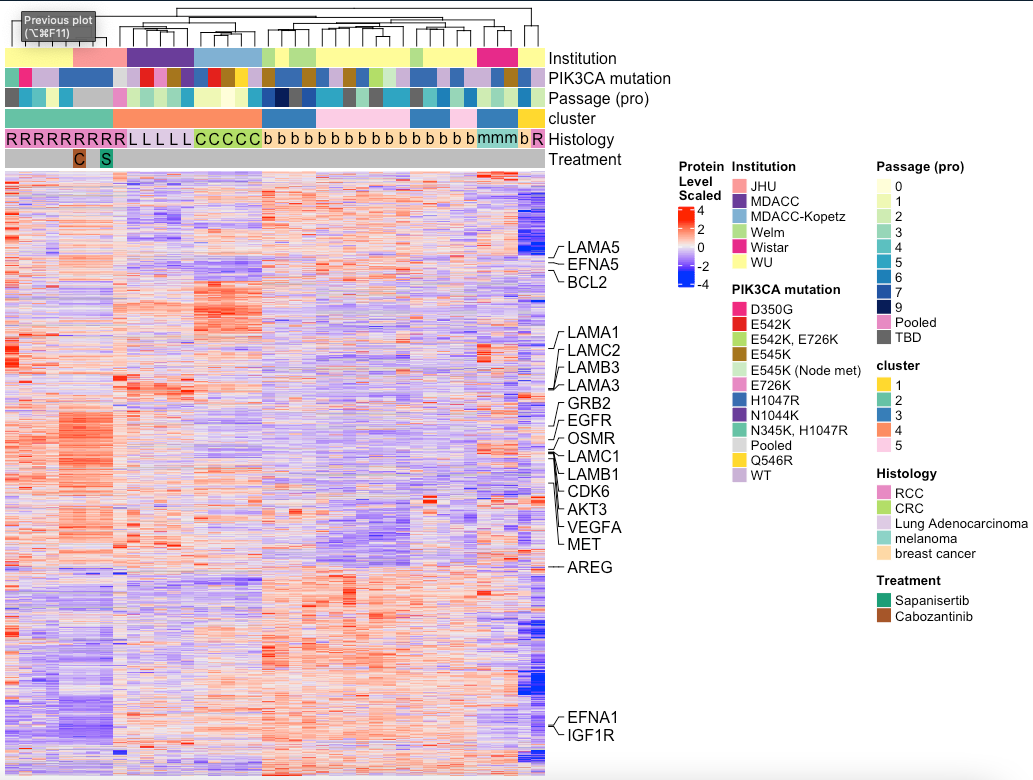
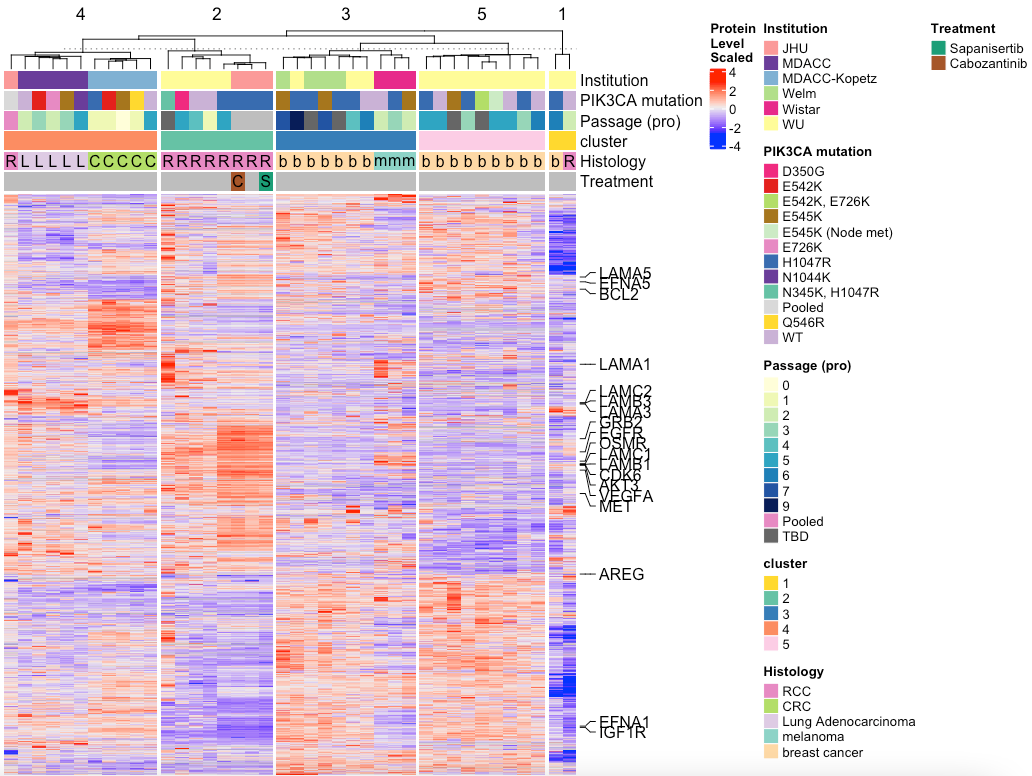
MakeTopProHeatmap(pancan_obj, palette_list = palette_list_pancan, split_column_by = 'cluster',
anno_pch_cols = c('Histology', "Treatment"),
anno_regular = c("Institution","PIK3CA mutation","Passage (pro)", 'cluster'))
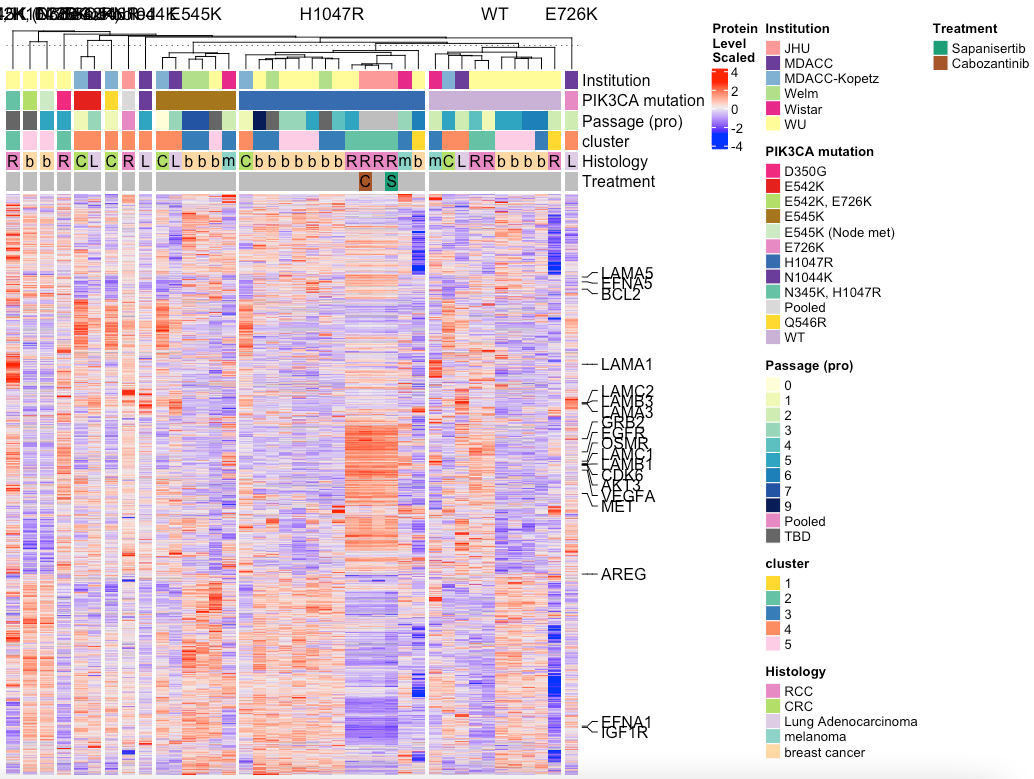
# Split by mutation
MakeTopProHeatmap(pancan_obj, palette_list = palette_list_pancan, split_column_by = 'PIK3CA mutation',
anno_pch_cols = c('Histology', "Treatment"),
anno_regular = c("Institution","PIK3CA mutation","Passage (pro)", 'cluster'))
Individual Cancer Type
### INdividual
## RCC
obj_list['RCC'](/simoncmo/pik3ca_proteomics/wiki/'RCC') = MakeDataObj(protein_mtx_human, meta, histology = 'RCC') %>% MakeCluster(k=4) %>% MakeUmap
dir.create('figure/RCC/')
pdf('figure/RCC/Heatmap.pdf', width = 10, height = 10)
MakeTopProHeatmap(obj_list['RCC'](/simoncmo/pik3ca_proteomics/wiki/'RCC'), palette_list = palette_list_pancan, split_column = F)
MakeTopProHeatmap(obj_list['RCC'](/simoncmo/pik3ca_proteomics/wiki/'RCC'), palette_list = palette_list_pancan, split_column_by = 'PIK3CA mutation')
MakeTopProHeatmap(obj_list['RCC'](/simoncmo/pik3ca_proteomics/wiki/'RCC'), palette_list = palette_list_pancan, split_column_by = 'cluster')
dev.off()
pdf('figure/RCC/Umap.pdf', width = 10, height = 10)
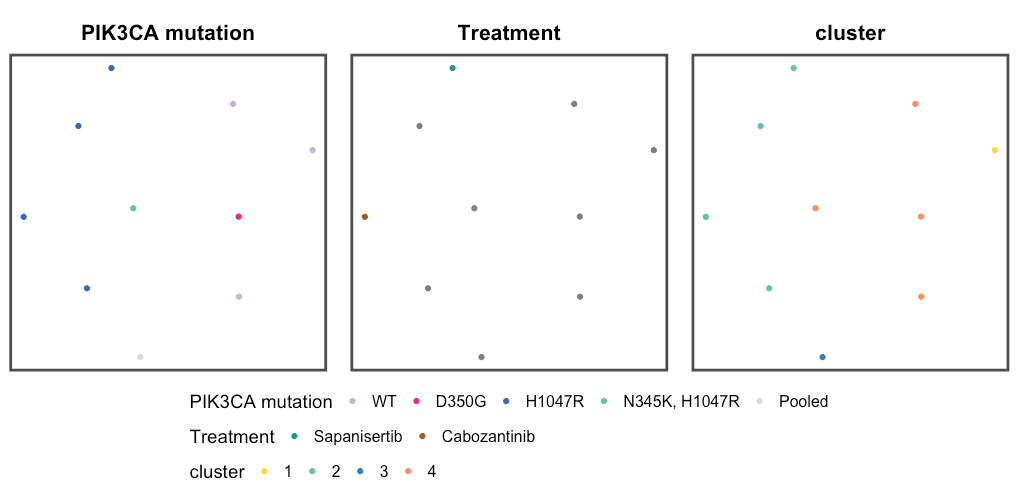
MakeMultipleDimPlot(obj_list['RCC'](/simoncmo/pik3ca_proteomics/wiki/'RCC'), palette_list = palette_list_pancan, column_names = c('PIK3CA mutation','Treatment','cluster'))
dev.off()