Use R to save table into Excel - openmpp/openmpp.github.io GitHub Wiki
It is a convenient to use GNU R to prepare model parameters and analyze output values. There are two different R APIs which we can use for openM++ models:
- openMpp package: simple and convenient specially for desktop users, upstream and downstream analysis;
-
omsJSON web-service API: preferable choice to run models on computational clusters and in cloud.
There is also an excelent R package created by Matthew T. Warkentin available at: mattwarkentin.github.io/openmpp.
Below is an example how to use oms JSON web-service to read multiple output table values from multiple model runs and save it into XLSX file:
- using
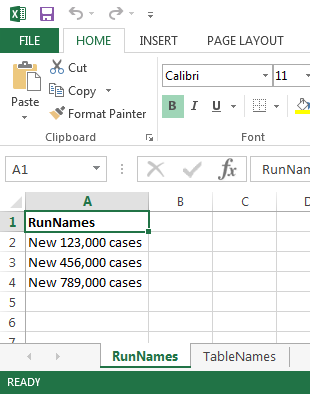
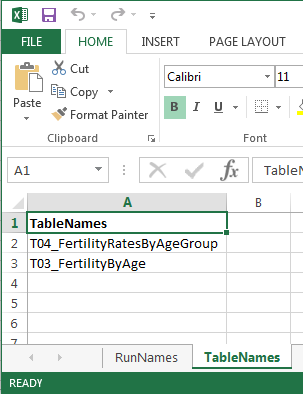
RiskPathsdemo model - reading model run names and output table names from input Excel file as on screenshots below
- for each table retriving output values for all model runs
- retrieving model runs metadata: run name, description, notes, date and time
- retrieving output tables metadata: name, description and notes
- saving each table output values as separate Excel workbook sheet
- saving all model runs metadata and tables metadata as separate sheets
Input Excel workbook:
Output Excel workbook:
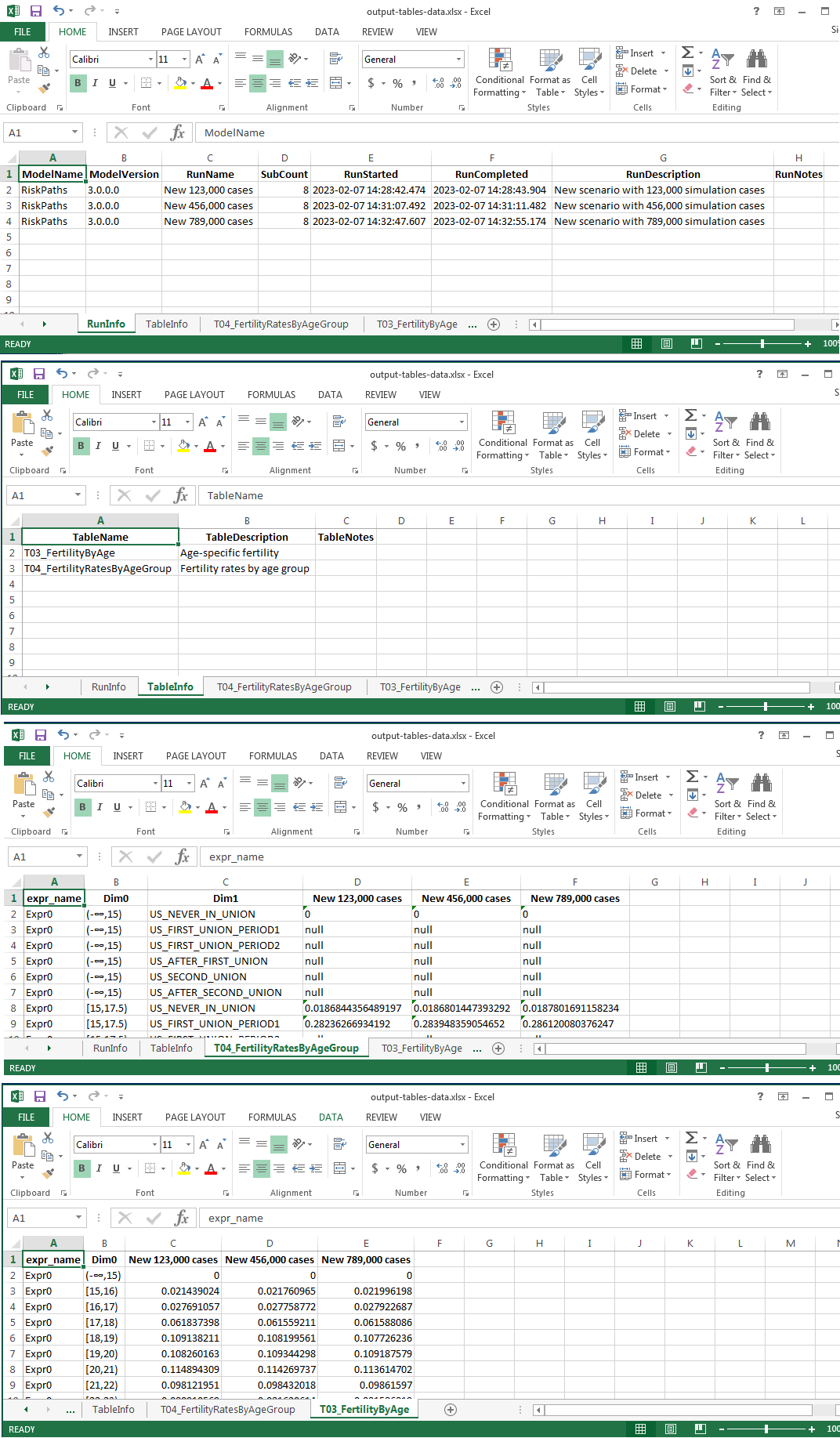
#
# Read multiple tables from multiple model runs and save it as XLSX file
# Also save model runs metadata and tables metadata (name, description, notes) into .csv files
# Model run names and table names are coming from another input XLSX file
#
# If any of library below is not installed then do:
# install.packages("jsonlite")
# install.packages("httr")
# install.packages("readxl")
# install.packages("writexl")
#
library("jsonlite")
library("httr")
library("readxl")
library("writexl")
# Include openM++ helper functions from your $HOME directory
#
source("~/omsCommon.R")
#
# Model digest of RiskPaths version 3.0.0.0: "d90e1e9a49a06d972ecf1d50e684c62b"
# We MUST use model digest if there are multiple versions of the model published.
# We can use model name if only single version of the model is published.
#
md <- "d90e1e9a49a06d972ecf1d50e684c62b"
# oms web-service URL from file: ~/oms_url.txt
#
apiUrl <- getOmsApiUrl()
# model runs can be identified by digest, by run stamp or by run name
# run digest is unique and it preferable way to identify model run
# run names are user friendly may not be unique
#
# read model run names from some XLSX file,
# it must have sheet name = "RunNames" with A column "RunNames"
#
rn <- read_xlsx(
"model-runs-to-read-and-tables-to-read.xlsx",
sheet = "RunNames",
col_types = "text"
)
# read table names from some XLSX file,
# it must have sheet name = "TableNames" with A column "TableNames"
#
tn <- read_xlsx(
"model-runs-to-read-and-tables-to-read.xlsx",
sheet = "TableNames",
col_types = "text"
)
# get table information
#
rsp <- GET(paste0(
apiUrl, "model/", md, "/text"
))
if (http_type(rsp) != 'application/json') {
stop("Failed to get first model info")
}
jr <- content(rsp)
tTxt <- jr$TableTxt
tableInfo <- data.frame()
for (t in tTxt) {
for (tbl in tn$TableNames)
{
if (t$Table$Name == tbl) {
ti <- data.frame(
TableName = tbl,
TableDescription = t$TableDescr,
TableNotes = t$TableNote
)
tableInfo <- rbind(tableInfo, ti)
break
}
}
}
# get run information
#
runInfo <- data.frame()
for (run in rn$RunNames)
{
rsp <- GET(paste0(
apiUrl, "model/", md, "/run/", URLencode(run, reserved = TRUE), "/text"
))
if (http_type(rsp) != 'application/json') {
stop("Failed to get first run info of: ", run)
}
jr <- content(rsp)
ri <- data.frame(
ModelName = jr$ModelName,
ModelVersion = jr$ModelVersion,
RunName = jr$Name,
SubCount = jr$SubCount,
RunStarted = jr$CreateDateTime,
RunCompleted = jr$UpdateDateTime,
RunDescription = "",
RunNotes = ""
)
if (length(jr$Txt) > 0) {
ri$RunDescription <- jr$Txt[[1]]$Descr
ri$RunNotes <- jr$Txt[[1]]$Note
}
runInfo <- rbind(runInfo, ri)
}
# for each table do:
# combine all run results and write it into some .xlsx file
#
shts <- list(
RunInfo = runInfo,
TableInfo = tableInfo
)
for (tbl in tn$TableNames)
{
allCct <- NULL
isFirst <- TRUE
for (run in rn$RunNames)
{
cct <- read.csv(paste0(
apiUrl, "model/", md, "/run/", URLencode(run, reserved = TRUE), "/table/", tbl, "/expr/csv"
))
# build a pivot table data frame:
# use first run results to assign all dimensions and measure(s)
# from all subsequent model run bind only expr_value column
if (isFirst) {
allCct <- rbind(allCct, cct)
isFirst <- FALSE
} else {
cval <- data.frame(expr_value = cct$expr_value)
allCct <- cbind(allCct, cval)
}
# use run name for expression values column name
names(allCct)[names(allCct) == 'expr_value'] <- run
}
shts[[ tbl ]] <- allCct
}
write_xlsx(shts, paste0("output-tables-data.xlsx"))