Microdata Output - openmpp/openmpp.github.io GitHub Wiki
Home > Model Development Topics > Microdata Output
Microdata output allows a model to output records containing the values of selected entity attributes during a run for later use. This topic describes microdata output from a model developer perspective.
- Introduction
- Topic outline
- Quick start How to build and run a model with microdata output
- Worked example 1a Entity life cycle
- Worked example 1b Entity life cycle with event context
- Worked example 1c Entity life cycle with event filtering
- Worked example 2a Output using a hook to a model event
- Worked example 2b Output using a hook to a self-scheduling attribute
-
Worked example 2c Output by calling
write_microdatain model code - Worked example 3 Database output in a time-based model
- Worked example 4 Database output in a complex case-based model
- Microdata output modes Text mode and database mode
- Microdata output control Enabling microdata and microdata attributes
- Run-time settings Run-time settings
- Build-time settings Build-time settings
- Writing microdata from model code Controlling microdata output from model code
- The microdata key The purpose of the microdata key and how to set it
A model built with microdata output capability can output records containing the values of entity attributes. As well as attribute values, each microdata output record contains a microdata key to match corresponding records between runs.
By default, a model does not have microdata output capability. See Enabling microdata output or Quick start on how to build a model with microdata output capability.
Two microdata output modes are supported: text mode and database mode. Text mode is targeted more to model developers, while database mode is targeted more to users of production models and to future OpenM++ run-time tabulation functionality. Both modes can be active in the same run.
Text mode writes microdata to text files in csv format.
Text mode can filter output at run using event context.
Text mode output can include an additional column showing the event context of the record.
Database mode writes microdata to the model database, from which it can be extracted using dbcopy or an API.
Database mode will be used for future OpenM++ run-time tabulation functionality, including microdata comparisons between runs.
Microdata output is controlled by run-time settings, build-time settings, and model code.
Run-time settings specify which attributes are output during a run, provided the model was built with microdata output capability. All attributes are available for selection at run-time without rebuilding the model. Some run-time settings apply only to text mode. Those text mode settings can filter records by event context and can create an additional column showing the event context for each record.
Build-time settings are statements in model code which make the model capable of microdata output and control related warning messages. Build-time settings can also (optionally) determine when microdata output is written in the entity life cycle: on entrance, on exit, or on the occurrence of an event.
Model code can write microdata explicitly by calling the supplied entity member function write_microdata.
The write_microdata function can be hooked to an existing entity function such as the implementation function of an event.
Quick Start shows how to build a model capable of microdata output and how to activate that capability in a model run.
The quick start is followed by several worked examples with illustrative inputs and outputs, mostly using the RiskPaths model.
The first group of examples
entity life cycle,
entity life cycle with event context, and
entity life cycle with event filtering
illustrate how to probe the life cycle of entities using microdata text mode.
A second group of examples
output using a hook to a model event,
output using a hook to a self-scheduling attribute, and
output by calling write_microdata in model code
illustrate how to control when microdata output occurs using model code.
The next example illustrates
database output in a time-based model.
The example outputs microdata for all entities in the IDMM model at the end of the run.
A Base run and a Variant are performed, and the results compared at the microdata level using files exported by sbcopy.
The final example illustrates
database output in a case-based model.
The example outputs microdata for all entities in the complex case-based model OncoSim.
A Base run and a Variant are performed, and summary microdata indicators (years lived and health system cost) are output for each Person at the end of each case.
The results are exported by dbcopy and analyzed to identify all cases which differed due to the parameter change in the Variant run, and by how much.
The worked examples are followed by subtopics which explore specifics in more detail:
- Microdata output modes
- Enabling microdata output
- Run-time settings
- Build-time settings
- Writing microdata from model code
- The microdata key
This subtopic contains the following sections.
- 1. Build model with microdata output capability
- 2. Create model
inifile with microdata output options - 3. Run model using microdata output
Add the following statements to the model source code file RiskPaths/code/ompp_framework.ompp:
options microdata_output = on;
options microdata_write_on_exit = on;Build the Release version of RiskPaths.
In Windows, the model executable will be RiskPaths/ompp/bin/RiskPaths.exe.
In Linux, the model executable will be RiskPaths/ompp-linux/bin/RiskPaths.
[back to quick start]
[back to topic contents]
In the same folder as the RiskPaths executable there may already be a copy of the default model ini file RiskPaths.ini.
If not create it using your IDE or a text editor such as Notepad.
Edit RiskPaths.ini to have the following content:
[Parameter]
SimulationCases = 5
[Microdata]
ToCsv = yes
Person = age, union_status, parity_status[back to quick start]
[back to topic contents]
Launch the model in its bin directory using the ini file created in the previous step.
RiskPaths -ini RiskPaths.ini
In Windows you can run the Release version of RiskPaths from inside Visual Studio as follows:
-
Solution ConfigurationstoReleaseandSolution Platformstox64 -
Project Properties > Configuration Properties > Debugging > Command Argumentsto
-ini RiskPaths.ini -
Project Properties > Configuration Properties > Debugging > Working Directoryto$(TargetDir) - To launch the model, do
Debug > Start without debuggingor PressCtrl-F5.
When the model run completes, the file RiskPaths.Person.microdata.csv should be present in the model bin directory and look like this:
key,age,union_status,parity_status
1,100,2,1
2,100,2,1
3,100,2,1
4,100,0,1
5,100,0,1
or formatted as a table, like this:
| key | age | union_status | parity_status |
|---|---|---|---|
| 1 | 100 | 2 | 1 |
| 2 | 100 | 2 | 1 |
| 3 | 100 | 2 | 1 |
| 4 | 100 | 0 | 1 |
| 5 | 100 | 0 | 1 |
The run-time settings output the attributes age, union_status, and parity_status. The leading column key can be used to match microdata records between runs.
The build-time option microdata_write_on_exit causes a microdata record to be written whenever an entity leaves the simulation. In RiskPaths there is no mortality and Person entities exit the simulation at age 100. The values of union_status and parity_status are those at that age, for each Person entity in the run.
The model log contains the following warning, which is expected.
Warning : model can expose microdata at run-time with output_microdata = on
[back to quick start]
[back to topic contents]
This example is the first of three which probe entity life cycle using microdata output in text mode. It continues the quick start example to output multiple microdata records for a single entity: when it enters the simulation, at each event, and when it leaves the simulation.
In ompp_framework.ompp, change the build-time microdata settings to
options microdata_output = on;
options microdata_write_on_enter = on;
options microdata_write_on_exit = on;
options microdata_write_on_event = on;Change the run-time settings in RiskPaths.ini to consist of only one case
[Parameter]
SimulationCases = 1
[Microdata]
ToCsv = yes
Person = age, union_status, parity_status
and run the model.
Here's the resulting microdata output in RiskPaths.Person.microdata.csv, with some rows elided.
| key | age | union_status | parity_status |
|---|---|---|---|
| 1 | 0 | 0 | 0 |
| 1 | 1 | 0 | 0 |
| 1 | 2 | 0 | 0 |
| 1 | 3 | 0 | 0 |
| ... | ... | ... | ... |
| 1 | 22.5 | 0 | 0 |
| 1 | 23 | 0 | 0 |
| 1 | 24 | 0 | 0 |
| 1 | 24.2609992115357 | 1 | 0 |
| 1 | 25 | 1 | 0 |
| 1 | 25.2609992115357 | 1 | 0 |
| 1 | 26 | 1 | 0 |
| 1 | 26.5378127283906 | 1 | 1 |
| 1 | 26.5378127283906 | 1 | 1 |
| 1 | 27 | 1 | 1 |
| 1 | 27.2609992115357 | 1 | 1 |
| 1 | 27.2609992115357 | 2 | 1 |
| 1 | 27.5 | 2 | 1 |
| 1 | 28 | 2 | 1 |
| 1 | 29 | 2 | 1 |
| 1 | 29.2609992115357 | 2 | 1 |
| 1 | 30 | 2 | 1 |
| ... | ... | ... | ... |
| 1 | 99 | 2 | 1 |
| 1 | 100 | 2 | 1 |
| 1 | 100 | 2 | 1 |
| 1 | 100 | 2 | 1 |
The microdata output shows the values of the attributes at every event in the life cycle. Multiple microdata records can occur at the same age due to multiple tied events at that age.
This example is the second of three which probe entity life cycle using microdata output in text mode. It continues the previous example, adding event context information to each microdata record.
Leave the build-time microdata settings in ompp_framework.ompp unchanged from the previous example:
options microdata_output = on;
options microdata_write_on_enter = on;
options microdata_write_on_exit = on;
options microdata_write_on_event = on;Activate the CsvEventColumn option by modifying the run-time settings in RiskPaths.ini so that it looks like this:
[Parameter]
SimulationCases = 1
[Microdata]
ToCsv = yes
CsvEventColumn = true
Person = age, union_status, parity_status
Run the model.
Here's the resulting microdata output in RiskPaths.Person.microdata.csv, with some rows elided.
| key | event | age | union_status | parity_status |
|---|---|---|---|---|
| 1 | (no event) | 0 | 0 | 0 |
| 1 | om_ss_event | 1 | 0 | 0 |
| 1 | om_ss_event | 2 | 0 | 0 |
| 1 | om_ss_event | 3 | 0 | 0 |
| ... | ... | ... | ... | ... |
| 1 | om_ss_event | 22.5 | 0 | 0 |
| 1 | om_ss_event | 23 | 0 | 0 |
| 1 | om_ss_event | 24 | 0 | 0 |
| 1 | Union1FormationEvent | 24.2609992115357 | 1 | 0 |
| 1 | om_ss_event | 25 | 1 | 0 |
| 1 | om_ss_event | 25.2609992115357 | 1 | 0 |
| 1 | om_ss_event | 26 | 1 | 0 |
| 1 | FirstPregEvent | 26.5378127283906 | 1 | 1 |
| 1 | om_ss_event | 26.5378127283906 | 1 | 1 |
| 1 | om_ss_event | 27 | 1 | 1 |
| 1 | om_ss_event | 27.2609992115357 | 1 | 1 |
| 1 | UnionPeriod2Event | 27.2609992115357 | 2 | 1 |
| 1 | om_ss_event | 27.5 | 2 | 1 |
| 1 | om_ss_event | 28 | 2 | 1 |
| 1 | om_ss_event | 29 | 2 | 1 |
| 1 | om_ss_event | 29.2609992115357 | 2 | 1 |
| 1 | om_ss_event | 30 | 2 | 1 |
| ... | ... | ... | ... | ... |
| 1 | om_ss_event | 99 | 2 | 1 |
| 1 | om_ss_event | 100 | 2 | 1 |
| 1 | DeathEvent | 100 | 2 | 1 |
| 1 | DeathEvent | 100 | 2 | 1 |
The microdata output now contains an event column showing the name of the event being implemented when each microdata record was output.
There is no event at the beginning of a case in a case-based model like RiskPaths,
so when the first entity in the case enters the simulation (no event) is shown in the event column.
If the event associated with microdata output is a self-scheduling event,
om_ss_event is shown in the event column. The internal self-scheduling event for an entity implements all self-scheduling attributes in the entity.
Note that
Event Trace
can be used to obtain more information about events, including the names of self-scheduling events.
The final three microdata output records all occur at age 100. Here's a detailed explanation of each of these apparent duplicate records:
The first is from the self-scheduling event which maintains the derived attribute self_scheduling_int(age). That derived attribute is in turn used in the declaration of the identity attribute integer_age:
actor Person //EN Individual
{
//EN Current integer age
LIFE integer_age = COERCE( LIFE, self_scheduling_int(age) );
...The second is from the event DeathEvent which is triggered by model logic and the ProbMort parameter immediately when integer_age is 100:
TIME Person::timeDeathEvent()
{
TIME event_time = TIME_INFINITE;
if (CanDie)
{
if (ProbMort[integer_age] >= 1)
{
event_time = WAIT(0);
}
...The third occurs when the entity leaves the simulation, because the option microdata_write_on_exit is on in the example. The event DeathEvent was the active event when the entity left the simulation, so that's what's shown in the event column.
Although it's not illustrated in this example, the name in the event column can be prefixed by a *.
This indicates that the active event is in a different entity than the one being output.
This can occur in a time-based model or in a case-based model with multiple entities in a case.
For example a ChildBirth event in a Person entity could cause a new Person entity to enter the simulation and generate a microdata output record.
The microdata record for the newborn would contain *ChildBirth in the event column to indicate that the active event was in a different entity than the microdata record.
This example is the third of three which probe entity life cycle using microdata output in text mode. It extends the previous example by filtering on specific events.
Leave the build-time microdata settings in ompp_framework.ompp unchanged from the previous example:
options microdata_output = on;
options microdata_write_on_enter = on;
options microdata_write_on_exit = on;
options microdata_write_on_event = on;Modify the run-time settings in RiskPaths.ini to increase the number of cases to 5000, and restrict output to two named events using the Events option:
[Parameter]
SimulationCases = 5000
[Microdata]
ToCsv = yes
CsvEventColumn = true
Person = age, union_status, parity_status
Events = Union1FormationEvent, FirstPregEvent
Run the model.
The resulting microdata output RiskPaths.Person.microdata.csv has 8,128 records and looks like this:
| key | event | age | union_status | parity_status |
|---|---|---|---|---|
| 1 | Union1FormationEvent | 24.2609992115357 | 1 | 0 |
| 1 | FirstPregEvent | 26.5378127283906 | 1 | 1 |
| 2 | Union1FormationEvent | 22.0523726276488 | 1 | 0 |
| 2 | FirstPregEvent | 24.6780778011483 | 1 | 1 |
| 3 | Union1FormationEvent | 17.050111243303 | 1 | 0 |
| 3 | FirstPregEvent | 20.024664717724 | 1 | 1 |
| 4 | FirstPregEvent | 17.4107170399441 | 0 | 1 |
| 5 | FirstPregEvent | 24.1577392012077 | 0 | 1 |
| 6 | Union1FormationEvent | 22.502915072767 | 1 | 0 |
| 6 | FirstPregEvent | 24.7534475294375 | 1 | 1 |
| ... | ... | ... | ... | ... |
This csv file can be used to perform multivariate statistical analysis.
For example, the csv file can be opened in Excel, filtered to just FirstPregEvent and a histogram generated to visualize the first birth distribution by age:
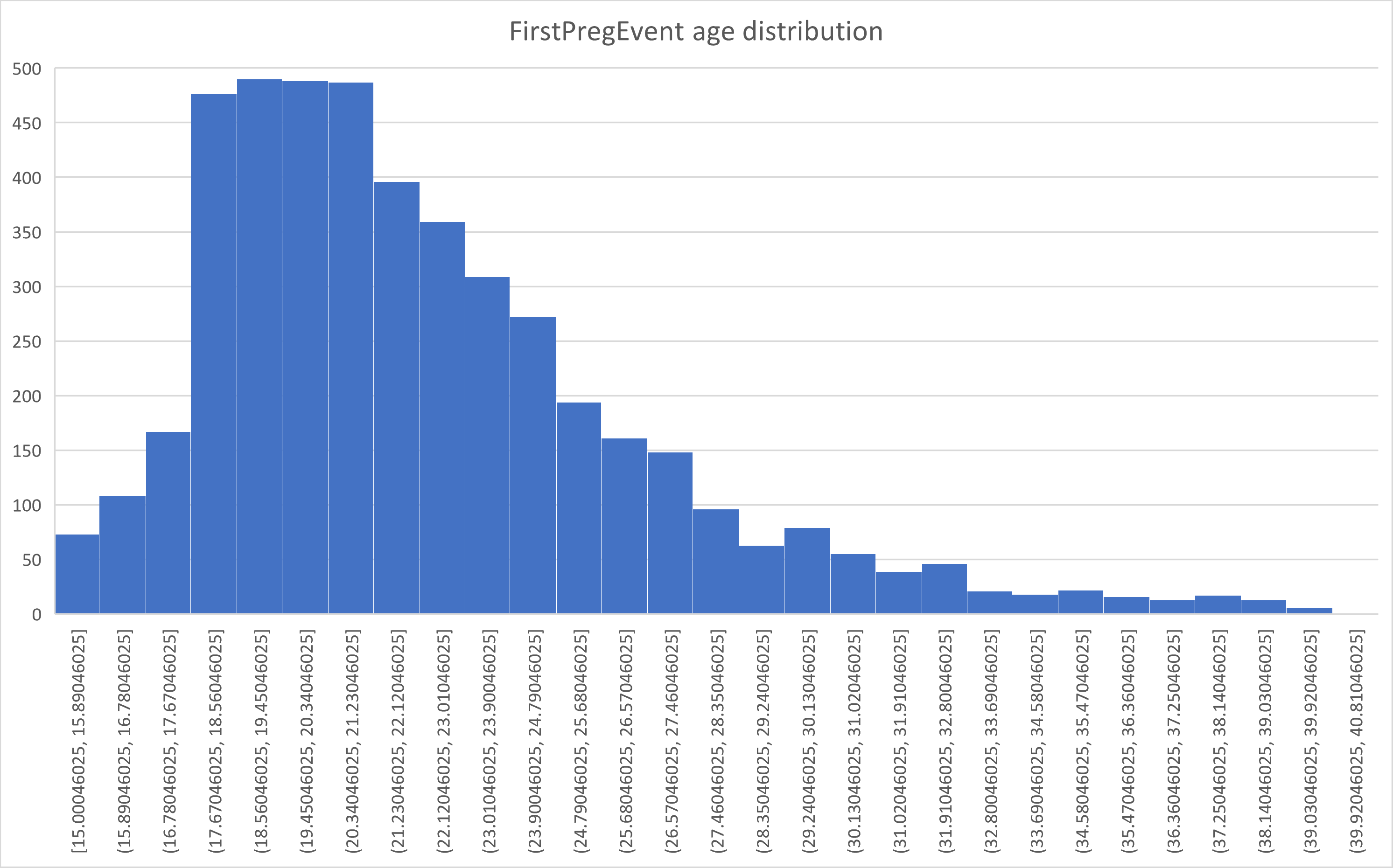
The data could be additionally filtered in Excel using the union_status column to visualize how union status affects the age distribution to produce the overall pattern.
This example is the first of three illustrating control of microdata output at build time using model code.
It outputs microdata whenever a specific event occurs using a hook in model code,
specifically whenever FirstPregEvent occurs in RiskPaths.
In RiskPaths, prepare the event implement function for hooks by adding the required statement at the end of the event implement function FirstPregEvent:
void Person::FirstPregEvent()
{
parity_status = PS_PREGNANT;
hook_FirstPregEvent();
}Next, add code to hook the built-in function write_microdata to FirstPregEvent:
actor Person {
hook write_microdata, FirstPregEvent;
};In ompp_framework.ompp, turn off options which automatically write microdata, which were previously turned on in example 1.
//options microdata_write_on_enter = on;
//options microdata_write_on_exit = on;
//options microdata_write_on_event = on;The statements inserted in example 1 were commented to revert to the default value off.
This means that only explicit calls to write_microdata will generate microdata output.
Set the number of cases to 20 in RiskPaths.ini:
[Parameter]
SimulationCases = 20
[Microdata]
ToCsv = yes
Person = age, union_status, parity_statusRun the model.
The microdata output file RiskPaths.Person.microdata.csv should look like this:
| key | age | union_status | parity_status |
|---|---|---|---|
| 1 | 26.5378127283906 | 1 | 1 |
| 2 | 24.6780778011483 | 1 | 1 |
| 3 | 20.024664717724 | 1 | 1 |
| 4 | 17.4107170399441 | 0 | 1 |
| 5 | 24.1577392012077 | 0 | 1 |
| 6 | 24.7534475294375 | 1 | 1 |
| 7 | 18.2797585879836 | 1 | 1 |
| 8 | 22.110326319997 | 1 | 1 |
| 9 | 21.2430736420085 | 1 | 1 |
| 10 | 29.168835553187 | 1 | 1 |
| 12 | 37.7955780112222 | 2 | 1 |
| 14 | 26.9550960057145 | 1 | 1 |
| 15 | 21.6012847802494 | 0 | 1 |
| 16 | 20.3178392448776 | 1 | 1 |
| 18 | 22.8298415328563 | 1 | 1 |
| 19 | 26.7999269606788 | 1 | 1 |
| 20 | 19.0257883348614 | 1 | 1 |
The microdata file shows the values of attributes at all occurrences of the FirstPregEvent in the run.
It could, for example, be used to chart the distribution of age at first birth using a downstream application like Excel or R, similar to example 1c.
This example is the second of three illustrating control of microdata output at build time using model code.
It outputs microdata records giving a snapshot of an entity at each integer age, using a hook to a self-scheduling attribute.
Change the hook in the previous example to
actor Person {
hook write_microdata, self_scheduling_int(age);
};and simulate a single case by modifying RiskPaths.ini:
[Parameter]
SimulationCases = 1
[Microdata]
ToCsv = yes
Person = age, union_status, parity_statusRun the model. Microdata output should look like this:
| key | age | union_status | parity_status |
|---|---|---|---|
| 1 | 1 | 0 | 0 |
| 1 | 2 | 0 | 0 |
| 1 | 3 | 0 | 0 |
| 1 | 4 | 0 | 0 |
| ... | ... | ... | ... |
| 1 | 26 | 1 | 0 |
| 1 | 27 | 1 | 1 |
| 1 | 28 | 2 | 1 |
| 1 | 29 | 2 | 1 |
| ... | ... | ... | ... |
| 1 | 100 | 2 | 1 |
The microdata output contains a snapshot of the attributes at each integer age.
The technique of hooking write_microdata to a self-scheduling or a trigger attribute will not output microdata if the current event causes the entity to exit the simulation.
That's because once the entity has exited the simulation no further events occur in it,
including the internal self-scheduling event to which write_microdata is hooked.
This example is the third of three illustrating control of microdata output at build time using model code.
It outputs microdata directly by calling the entity function write_microdata explicitly in model code.
Remove any changes to RiskPaths model code made in previous examples.
In ompp_framework.ompp, insert the single statement
options microdata_output = on;Insert a call to write_microdata in the implementation function of the FirstPreg event in the module Fertility.mpp:
void Person::FirstPregEvent()
{
parity_status = PS_PREGNANT;
write_microdata();
}Set the run-time settings in RiskPaths.ini as follows:
[Parameter]
SimulationCases = 5
[Microdata]
ToCsv = yes
Person = age, union_status, parity_status
Run the model.
Output should look as follows:
key,age,union_status,parity_status
1,26.5378127283906,1,1
2,24.6780778011483,1,1
3,20.024664717724,1,1
4,17.4107170399441,0,1
5,24.1577392012077,0,1
This example could be accomplished without using a direct call to write_microdata.
In a more complex model, a call to write_microdata could be placed inside conditional model logic,
for example to output microdata when a rare causative path is taken in model logic, to probe correctness.
This example outputs microdata in database mode for the time-based model IDMM.
Two runs Base and Variant are performed with an incremental parameter change.
Microdata with infection status is output for all Host entities at the end of the run.
Each run consists of multiple replicates.
The dbcopy utility is used to extract the microdata for the two runs.
Excel is used to import the microdata and construct a table showing the concordance of disease state at the microdata level between the Base and Variant runs.
Modify the IDMM model to activate microdata output when entities leave the simulation by adding the following statements to ompp_framework.ompp:
options microdata_output = on;
options microdata_write_on_exit = on;Rebuild the model.
Arrange that IDMM uses the file IDMM.ini to get run-time settings (see quick start), and set the contents of IDMM.ini to create a run named Base as follows:
[OpenM]
SubValues = 5
Threads = 5
RunName = Base
[Parameter]
NumberOfHosts = 10000
ImmunePhaseDuration = 20.0
[Microdata]
ToDb = yes
Host = disease_phaseThese settings create a Base run with 5 replicates, each with a population of 10,000 Host entities.
Run the model.
The log file should contain a line like
2023-01-13 17:01:04.874 Warning : model can expose microdata at run-time with output_microdata = on
which indicates that the version of IDMM is capable of writing microdata. It should also contain a line similar to
2023-01-13 17:01:08.295 Writing microdata into database, run: 103
which indicates that the model is merging microdata from replicates into the database when the run completes.
Change the file IDMM.ini, modifying RunName and ImmunePhaseDuration for a second run named Variant:
[OpenM]
SubValues = 5
Threads = 5
RunName = Variant
[Parameter]
NumberOfHosts = 10000
ImmunePhaseDuration = 22.0
[Microdata]
ToDb = yes
Host = disease_phaseThe Variant run is the same as the Base run, except for a 10% increase in the duration of protective immunity from a previous infection.
Run the model.
The model database now contains results for the two runs Base and Variant.
Open a command shell.
Change the current directory to the ompp/bin directory of the IDMM model.
Run dbcopy to extract the microdata results from the model database to csv files using the command
dbcopy -dbcopy.To csv -dbcopy.ModelName IDMM
By default dbcopy looks for a model database in the current directory, so it's not necessary in this example to provide it the path of the model database.
Console output should be similar to the following:
C:\Development\X\ompp\models\IDMM\ompp\bin>%OM_ROOT%\bin\dbcopy -dbcopy.To csv -dbcopy.ModelName IDMM
2023-01-13 17:01:45.580 Model IDMM
2023-01-13 17:01:45.599 Model run 102 Base
2023-01-13 17:01:45.600 Parameters: 13
2023-01-13 17:01:45.609 Tables: 3
2023-01-13 17:01:45.622 Microdata: 1
2023-01-13 17:01:45.688 Model run 103 Variant
2023-01-13 17:01:45.690 Parameters: 13
2023-01-13 17:01:45.700 Tables: 3
2023-01-13 17:01:45.712 Microdata: 1
2023-01-13 17:01:45.781 Workset 101 Default
2023-01-13 17:01:45.782 Parameters: 13
2023-01-13 17:01:45.798 Done.
The console output above was done on Windows.
There would be minor cosmetic differences in Linux.
Note the use of the global environment variable OM_ROOT to ensure that the version of dbcopy matches the version of OpenM++ used to build the model.
The dbcopy log output shows the extraction of the microdata for the two runs Base and Variant.
dbcopy creates a folder IDMM The folder structure of dbcopy output looks like
C:\OMPP\MODELS\IDMM\OMPP\BIN\IDMM
├───run.Base
│ ├───microdata
│ ├───output-tables
│ └───parameters
├───run.Variant
│ ├───microdata
│ ├───output-tables
│ └───parameters
└───set.Default
Each microdata sub-folder contains a file named Host.csv containing the microdata of Host entities for the run. Had microdata for the Ticker actor been requested in the run, a file Ticker.csv would also be present. The first few records of IDMM/run.Base/microdata/Host.csv look like this:
| key | disease_phase |
|---|---|
| 10 | DP_SUSCEPTIBLE |
| 11 | DP_LATENT |
| 12 | DP_IMMUNE |
| 13 | DP_LATENT |
| 14 | DP_IMMUNE |
| 15 | DP_LATENT |
| 16 | DP_SUSCEPTIBLE |
| 17 | DP_LATENT |
| 18 | DP_SUSCEPTIBLE |
| 19 | DP_IMMUNE |
| 20 | DP_IMMUNE |
For large output files, one can use the dbcopy option -dbcopy.IdCsv to output numeric id's instead of alphanumeric codes.
The default microdata key entity_id is used in this example.
entity_id is unique for all entities in a run, and will correspond to the same entity in two IDMM runs provided the runs have the same number of entities per replicate and the same number of replicates.
The two files run.Base/microdata/Host.csv and run.Variant/microdata/Host.csv were imported to Excel, and the 50,000 rows matched one-to-one.
Below is an Excel PivotTable (aka cross-tab) which counts the 50,000 Host entities at the end of the runs, classified by disease phase in the Base run (rows) and disease phase in the Variant run (columns).
| Base↓/Variant→ | DP_IMMUNE | DP_INFECTIOUS | DP_LATENT | DP_SUSCEPTIBLE | All |
|---|---|---|---|---|---|
| DP_IMMUNE | 24284 | 806 | 390 | 2649 | 28129 |
| DP_INFECTIOUS | 1849 | 137 | 92 | 334 | 2412 |
| DP_LATENT | 2268 | 94 | 67 | 354 | 2783 |
| DP_SUSCEPTIBLE | 13932 | 421 | 352 | 1971 | 16676 |
| All | 42333 | 1458 | 901 | 5308 | 50000 |
The lexicographic ordering of disease phase in the table does not follow the ordering in model code, which makes the table harder to interpret.
The intuitive order is Susceptible, Latent, Infectious, Immune.
That could be addressed by revising the DISEASE_PHASE classification codes in IDMM model code to align lexicographic order with model code order, e.g.
classification DISEASE_PHASE //EN Disease phase
{
//EN Susceptible
DP0_SUSCEPTIBLE,
//EN Latent
DP1_LATENT,
//EN Infectious
DP2_INFECTIOUS,
//EN Immune
DP3_IMMUNE
};Alternatively, the microdata could have been exported using the option -dbCopy.IdCsv to output 0,1,2,3 instead of codes in the csv files.
However, numeric id's in table rows and columns are not informative.
From the table, the level of coherence between Base and Variant at the end of the simulations is not high.
This could be because
- a 10% increase in the duration of immunity is not as minor as one might think a priori;
- the increase in duration of immunity is expected to increase the period of epidemic cycles, which would cause epidemic cycles to be out of phase between
BaseandVariantat the end of the simulations; -
IDMMsimulates a highly interacting population which can diverge rapidly from a small initial perturbation; - simulation divergence is accelerated because
IDMMdoes not use entity-specific random number generators for decoherence control.
This example illustrates run comparison at the microdata level using a large scale complex case-based model (a working version of the Statistics Canada OncoSimX model).
This example is divided into the following sections:
The default microdata key entity_id is not suitable for run comparison in OncoSimX, so a model-specific definition of get_microdata_key was added to model code.
A pair of attributes (years lived and health system cost) were output for each Person entity at the end of each case.
A Base run with 500,000 cases and 12 replicates was performed with microdata output enabled, in database mode.
A Variant run was performed, changing a single scalar parameter.
Results for both runs were exported using dbcopy to csv files and analyzed in Excel to identify all cases which differed between Base and Variant runs for either of the two attributes.
The mechanical steps in this example are similar to those in the previous example.
[back to example 4 sections]
[back to topic contents]
The model code was modified to enable microdata output when the Person in each case exits the simulation by adding the following statements to model code.
options microdata_output = on;
options microdata_write_on_exit = on;In OncoSimX a case contains exactly one Person entity, but might contain other entities depending on the simulation, such as one or more Tumour entities.
Because the built-in attribute entity_id is incremented whenever a new entity is created,
entity_id is unsuitable as a microdata key to match corresponding Person entities between two OncoSimX runs.
However, the built-in attribute case_id is suitable as a microdata key for Person because it has a one-to-one relationship with the single Person entity in each case, and this relationship is robust across runs provided the runs have the same number of cases and replicates.
A function definition of Person::get_microdata_key was added to model code so that case_id is used as the microdata key for Person entities instead of entity_id:
uint64_t Person::get_microdata_key()
{
return case_id;
}[back to example 4 sections]
[back to topic contents]
The model was run using the settings file OncoSimX/ompp/bin/OncoSimX.ini, like previous examples.
The following run settings were used for the Base run:
[OpenM]
SubValues = 12
Threads = 12
RunName = Base
[Parameter]
SimulationSeed = 1
SimulationCases = 500000
MaxConsecutiveHpvTreatmentAllowed = 2
[Microdata]
ToDb = yes
Person = age, cancer_cost_allThe parameter MaxConsecutiveHpvTreatmentAllowed was chosen arbitrarily for this example.
A scalar parameter rather than an array parameter was chosen to make this example simpler, because the value of a scalar parameter can be specified in a model run ini file, obviating the need to set up and use a directory for Variant parameters which differ from Base.
Because the microdata for a Person entity is output when a Person leaves the simulation at death,
the attribute cancer_cost_all will contain lifetime cancer-related costs and
the age attribute will contain the duration of life in years.
These two attributes are measures of benefit and cost at the Person level.
The case_seed attribute can be useful to probe a case of interest in a subsequent run, but there is no need to include it in the Person microdata attributes because the key column already contains the value of case_seed, as described above.
For Variant, the parameter MaxConsecutiveHpvTreatmentAllowed was changed from 2 to 1, and RunName was changed to name the run Variant:
[OpenM]
SubValues = 12
Threads = 12
RunName = Variant
[Parameter]
SimulationSeed = 1
SimulationCases = 500000
MaxConsecutiveHpvTreatmentAllowed = 1
[Microdata]
ToDb = yes
Person = age, cancer_cost_all[back to example 4 sections]
[back to topic contents]
After the runs completed, microdata results were extracted from the database using dbcopy as in the
previous example.
Here's the Windows command session:
C:\Development\X\models\OncoSimX\ompp\bin>%OM_ROOT%\bin\dbcopy -dbcopy.To csv -dbcopy.ModelName OncoSimX
2023-01-14 18:00:10.259 Model OncoSimX
2023-01-14 18:00:10.392 Model run 102 Base
2023-01-14 18:00:10.392 Parameters: 402
2023-01-14 18:00:16.227 250 of 402: IncidenceRatesHpvMultiplier
2023-01-14 18:00:18.842 Tables: 27
2023-01-14 18:00:25.153 0 of 27: CervicalCancer_TreatmentCost_Table all accumulators
2023-01-14 18:00:31.685 1 of 27: Cervical_Cancer_Cases_PAY_Table all accumulators
2023-01-14 18:00:36.055 7 of 27: Cervical_Cancer_ICER_Table_Discounted all accumulators
2023-01-14 18:00:42.357 26 of 27: Hpv_Screening_Costs_Prov_Table all accumulators
2023-01-14 18:00:43.923 Microdata: 1
2023-01-14 18:00:45.023 Model run 103 Variant
2023-01-14 18:00:45.023 Parameters: 402
2023-01-14 18:00:51.051 250 of 402: IncidenceRatesHpvMultiplier
2023-01-14 18:00:53.794 Tables: 27
2023-01-14 18:01:00.062 0 of 27: CervicalCancer_TreatmentCost_Table all accumulators
2023-01-14 18:01:06.623 1 of 27: Cervical_Cancer_Cases_PAY_Table all accumulators
2023-01-14 18:01:11.144 8 of 27: Cervical_Cancer_LifetimeCost_Table
2023-01-14 18:01:17.385 26 of 27: Hpv_Screening_Costs_Prov_Table all accumulators
2023-01-14 18:01:18.976 Microdata: 1
2023-01-14 18:01:20.115 Workset 101 Default
2023-01-14 18:01:20.116 Parameters: 402
2023-01-14 18:01:26.157 250 of 402: IncidenceRatesHpvMultiplier
2023-01-14 18:01:29.024 Done.
The first rows of microdata output for the Base run in the file OncoSimX/ompp/bin/OncoSimX/run.Base/microdata/Person.csv look like this:
key,age,cancer_cost_all
0,79.4991129115706,45100.08867191
1,67.281040126587,2229.937944223
2,87.4865314659319,1670.3732276699
3,0.379665603266858,0
The first rows of microdata for the Variant run are identical.
However, some of the 500,000 microdata output records differ between Variant and Base.
[back to example 4 sections]
[back to topic contents]
An Excel workbook was created and used to
- load the
csvmicrodata forBaseandVariantas queries, renaming columns to distinguishBaseandVariant; - merge the two queries matching on
keyto create a new query with one row for each case andBaseandVariantmicrodata in distinct columns. - add a column to the merge query to compute the Variant-Base difference in years lived;
- add a column to the merge query to compute the Variant-Base difference in lifetime cancer-related costs;
- add a column named
Differsto compute whether a microdata record differed in either years lived or cost betweenBaseandVariant.
A dynamic filter was applied to the Differs column of the Excel table for the merge query to display all records which differed between Variant and Base.
13 of the 500,000 microdata records differed, as follows:
| key | life(base) | cost(base) | life(variant) | cost(variant) | life(delta) | cost(delta) | Differs |
|---|---|---|---|---|---|---|---|
| 26847 | 82.90 | 9,099 | 82.90 | 10,570 | 0.0000 | 1,471 | TRUE |
| 59368 | 89.07 | 60,812 | 89.07 | 61,528 | 0.0000 | 717 | TRUE |
| 208131 | 72.68 | 40,304 | 98.16 | 19,647 | 25.4839 | -20,657 | TRUE |
| 214559 | 94.60 | 31,285 | 94.60 | 27,932 | 0.0000 | -3,353 | TRUE |
| 229714 | 86.53 | 25,446 | 86.53 | 13,450 | 0.0000 | -11,996 | TRUE |
| 231202 | 95.18 | 101,255 | 95.18 | 100,388 | 0.0000 | -867 | TRUE |
| 247895 | 97.40 | 40,914 | 97.40 | 9,396 | 0.0000 | -31,518 | TRUE |
| 290098 | 92.17 | 13,059 | 92.17 | 14,461 | 0.0000 | 1,402 | TRUE |
| 302510 | 78.51 | 63,695 | 78.51 | 54,770 | 0.0000 | -8,926 | TRUE |
| 357201 | 78.91 | 8,080 | 78.91 | 9,482 | 0.0000 | 1,402 | TRUE |
| 436603 | 39.75 | 112,787 | 39.75 | 111,870 | 0.0000 | -916 | TRUE |
| 438020 | 65.36 | 84,806 | 63.36 | 80,545 | -2.0000 | -4,261 | TRUE |
| 447567 | 94.15 | 34,830 | 94.15 | 32,333 | 0.0000 | -2,498 | TRUE |
The key column contains the value of case_seed and could be used to re-simulate any (or all) of these differing cases using Event Trace to explore the different causative pathways taken in the Base and Variant runs, and how those different pathways affected Person attributes.
These differences suggest that it might be interesting to understand how the change in MaxConsecutiveHpvTreatmentAllowed from 2 to 1 resulted in
- an additional ~25 years of life for
case_id208131, - both positive and negative changes in health system costs for cases which experienced no change in years lived,
-
case_id438020 living an exact integer number of years 2.0000 less inVariantcompared toBase.
Quite possibly all these Base-Variant differences are explained by different but realistic causative pathways taken in the two runs. That could be verified by comparing the Base and Variant causative pathways for individual differing cases using Event Trace, perhaps by tracing all events, event times, and attribute changes in a differing case and examining differences in the Base and Variant event trace outputs.
This example illustrates how microdata differences between two runs can augment aggregate differences by drilling down to the detail underlying the aggregate differences. It also illustrates how microdata differences from a marginal change to a single model parameter can probe model logic and causative pathways and assist in model validation.
[back to example 4 sections]
[back to topic contents]
Two distinct modes are supported:
Database mode
and
Text mode.
- Targeted primarily for use of a production model, to drill down to underlying microdata or to compare two runs at a microdata level.
- uniqueness of key is required
- all microdata output, including from multiple instances and multiple threads, is merged into the model database.
- no run-time event filtering (but can be done in model code with build-time settings).
-
dbcopycan be used to extract microdata tocsvfiles, supports numeric id's or codes. -
omscan be used to extract microdata - will support future functionality for run-time tabulation, including microdata compare (winner-loser).
- Targeted primarily to probe a model during development, validation, and debugging
- uniqueness of key is not required
- to trace file or to entity-specific csv files
- runs using multiple instances have distinct csv files for each instance
- multiple threads in an instance share csv files.
- optional event context column
- optional event filtering
It is one file per process, all threads do write into the same file. As it is today file name can be:
(a) typical developer / desktop use case: single process, single model run:
ModelName.Entity.microdata.csv
(b) MPI cluster / cloud use case: multiple processes, single model run:
ModelName.Entity.07.microdata.csv
07 is an example of process rank, zero padded It is not limited to 00 - 99, it can be as large as cluster allow us to have, in ComputeCanada can be 5 digits
(c) modelling task run, for example from R or Python using single process:
ModelName.Entity.2022_12_31_22_33_44_981.microdata.csv
2022_12_31_22_33_44_981 is a model run timestamp, time when model run started. Because modelling task run include multiple model runs then each run creates it own microdata cvs file(s)
(d) = c + b: modeling task run in cloud with MPI cluster, it is possible from R on our CPAC cloud:
ModelName.Entity.2022_12_31_22_33_44_981.07.microdata.csv
This subtopic is divided into the following sections:
- Enabling microdata and controlling warnings
- Weight-enabled models
- Internal attributes
- Attributes with many enumerators
A model is capable of writing microdata if and only if model code contains the following statement:
options microdata_output = on;
A model with microdata capability will write the following warning to the log whenever it is run:
Warning : model can expose microdata at run-time with microdata_output = on
If this is not a concern, for example if the model generates entities synthetically, this warning can be disabled by the following statement:
options microdata_output_warning = off;[back to microdata output control sections]
[back to topic contents]
A weight-enabled model which is also microdata-enabled will write the following message to the log when run
Note : model is weight-enabled and microdata-enabled, include entity_weight in Microdata for downstream weighted operations
as a reminder that the attribute entity_weight needs to be included in microdata output for downstream weighted tabulation.
[back to microdata output control sections]
[back to topic contents]
Some internal entity attributes are created by the OpenM++ compiler. For example, the compiler creates an identity attribute to implement the filter of an entity table. These internal entity attributes are normally hidden. They can be made visible, including as microdata, using the following statement:
options all_attributes_visible = on;[back to microdata output control sections]
[back to topic contents]
Attributes whose type is an enumeration with a large number of enumerators may not be eligible as microdata.
For example,
the following code fragment declares the Person attribute id_tracker with type ID_LIST which has 5,000,001 possible values (enumerators):
range ID_LIST{0, 5000000};
actor Person
{
ID_LIST id_tracker; //EN Unique identifier of each actor
};If microdata output is enabled, the OpenM++ compiler will emit a warning like
PersonCore.mpp(254): warning - attribute 'id_tracker' has 5000001 enumerators making it ineligible as microdata - consider using int.
and the attribute id_tracker will not be available as microdata at runtime.
However, if id_tracker is instead declared to be of type int instead of type ID_LIST,
no warning will be issued and id_tracker,
with the same integer values assigned in model code,
will be available as microdata at runtime.
The maximum number of enumerators for an attribute (of type enumerator) to be eligible as microdata is 1,000,
but can be raised or lowered using the option microdata_max_enumerators.
For example,
options microdata_max_enumerators = 500;will restrict microdata attributes of enumeration type to those with 500 or fewer enumerators.
The threshold only applies to attributes declared with enumeration types like range.
It does not apply to attributes declared with non-enumeration types such as int, counter, big_counter, etc.
Attributes with large numbers of enumerators can cause performance degradation or instability in the microdata viewer and the microdata tabulator due to the large number of cells being manipulated and displayed.
[back to microdata output control sections]
[back to topic contents]
Run time settings are specified as options, either on the model executable command line or in a model run ini file.
In an ini file, microdata options are in the [Microdata] section.
On the command line, they are given like -Microdata.Person age.
The following table lists all microdata run-time settings with an example and a short description.
| Option | Example | Description |
|---|---|---|
| entity |
Person = ageGroup,sex,time
|
Store the named attributes for the specified entity kind, e.g. the attributes ageGroup, sex, and time, for Person entities. |
| entity | Person = All |
Store all non-internal attributes of Person entities. |
ToDb |
true |
Write microdata entity attributes into database. Important: each microdata entity must have a unique key. Default is false. |
ToCsv |
true |
Write microdata entity attributes and events (if enabled) into csv file(s). each microdata entity is written in its own file. Default is false. |
UseInternal |
true |
Store all non-internal attributes of all entities. NOT recommended for production, use for debug only. Default is false. |
CsvDir |
path/to/some/directory |
Directory where microdata csv file(s) are written, must be an existing directory. Default is the current directory. |
ToTrace |
true |
Write microdata entity(s) attributes and events (if enabled) to model Trace output. Trace must be enabled to produce any output. Default is false. |
Events |
Birth,Union,Death |
Write selected events into Trace or csv file. |
CsvEventColumn |
true |
If true then write event name into csv file. Default is false. |
For a complete example of a run ini file, including the [Microdata] section, see
OpenM++ ini-file run options.
Build-time settings which enable a model to output microdata are described in Enabling microdata output. Other build-time options can output microdata during the simulation of each entity. The available options are:
| Option | Default | Description |
|---|---|---|
microdata_write_on_enter |
off |
microdata is written when an entity enters the simulation, before any event occurs in the entity. |
microdata_write_on_exit |
off |
microdata is written when an entity exits the simulation. |
microdata_write_on_event |
off |
microdata is written after an event occurs in an entity. |
These options can be combined.
If none of these options are on no microdata will be written unless model code does so explicitly by calling or hooking the built-in function write_microdata.
Note that attributes of an entity can change due to events in other linked entities in a model with interacting entities.
So, even if microdata_write_on_event is on, changes in attributes of an entity can be absent from microdata output for that entity.
For example, in IDMM, if an infectious Host A infects Host B through A's social contacts, the event associated with the infection occurs in A and not in B.
If one wanted to output Host microdata at the moment of infection, one could do so by calling output_microdata explicitly in model code.
Under construction
Microdata can be written by calling the built-in entity function write_microdata() from model code, either directly or by using a hook statement.
If a model is not enabled for microdata, calls to write_microdata have no effect.
Modgen-specific: The Modgen build of a cross-compatible model inserts a do-nothing version of write_microdata() into the Modgen-generated C++ code.
This allows use of write_microdata in model code without producing C++ build errors in the Modgen build of a x-compatible model.
A key is a unique identifier used to match entities or microdata records across runs.
It is a 64-bit value of C++ type uint64_t.
The key for an entity is returned by the entity member function get_entity_key().
If this function is not defined in model code, the OpenM++ compiler will provide a definition which returns the value of the built-in attribute entity_id.
The entity key is described further
here.
The key for a microdata output record is produced by the entity member function get_microdata_key().
If this function is not defined in model code, the OpenM++ compiler will provide a definition which computes and returns the following value:
10000 * get_entity_key() + om_microdata_counterwhere om_microdata_counter is an internally-maintained counter of output microdata records for each individual entity.
This formula produces unique microdata keys because it combines the unique entity_id with an entity-specific counter of microdata records output for that entity.
Uniqueness is guaranteed provided that fewer than 10,000 microdata records are output for a single entity.
For example, if microdata is output at each event using the microdata_write_on_event option,
the default microdata key would be
entity_id |
Event | get_microdata_key() |
|---|---|---|
| 42 | first | 420001 |
| 42 | second | 420002 |
| 101 | first | 1010001 |
| 101 | second | 1010001 |
The OpenM++ compiler generates C++ code to create and maintain om_microdata_counter
only if the get_microdata_key() function has not been defined for that kind of entity in model code.
Note that the entity key from get_entity_key() can be used both to calculate the result of get_microdata_key() and to support
local random streams
in model architectures which use both.
Uniqueness of the microdata key is enforced for Database mode, but is not enforced for Text mode.
A model run-time error will occur if uniqueness of the microdata key is violated in Database mode, with log output similar to the following:
2023-01-24 12:22:27.202 Writing microdata into database, run: 102
2023-01-24 12:22:28.525 : UNIQUE constraint failed: Host_g732a1637.run_id, Host_g732a1637.entity_key
2023-01-24 12:22:28.528 Error at microdata: 2100, 100, 3
2023-01-24 12:22:28.559 DB error: UNIQUE constraint failed: Host_g732a1637.run_id, Host_g732a1637.entity_key
The line Error at microdata: 2100, 100, 3 indicates that the record {2100, 100,3} violated key uniqueness. The first value is the non-unique key value, which is 2100 in this example.
The following values are the other attributes of the microdata record with the non-unique key
The case-based model in Example 4 supplies a custom implementation of get_microdata_key() to correctly match Person microdata results between two runs, because in that model the number of secondary entities in a case can vary between two runs.
The time-based model in Example 3 uses the standard implementation of get_microdata_key() because there are no additions to the starting population of Host entities created at the beginning of a run,
and runs of equal size are being compared.
The following hypothetical definition of get_microdata_key() uses the helper function xz_crc64 to combine the value of get_entity_key() and report_time to create the microdata key.
xz_crc64 creates a 64-bit key using the crc-64 open source checksum (hash) algorithm, and can use one value or combine multiple values together using successive calls.
uint64_t Host::get_microdata_key()
{
uint64_t key64 = 0;
auto entity_key = get_entity_key();
key64 = xz_crc64((uint8_t*)&entity_key, sizeof(entity_key), key64);
key64 = xz_crc64((uint8_t*)&report_time, sizeof(report_time), key64);
return key64;
}This definition might be used in a model which outputs a microdata record for each entity at each report_time,
if the number of entities might vary from one run to another due to parameter differences (e.g. fertility).
For a model based on a microdata input file where each input record has a unique personal identification number person_id,
and in which only a single microdata record is output for each Person,
a suitable definition might look like:
uint64_t Person::get_microdata_key()
{
uint64_t key64;
key64 = person_id;
return key64;
}