What you need to know - okeyoallan/bioinformatics-internship GitHub Wiki
To Note
To get into bioinformatics, you need to;
- Have a background knowledge in biology
- Have knowledge in programming languages such as Python, R, Perl e.tc.
- Have good communication and writing skills
- Good problem solving skills
- Have knowledge in editing tools such as Jupiter Notebook
- Have knowledge in data structures and various biological databases
- Have an idea about biological computational packages such as R, Matlab, among others
Biological databases
These are libraries of biological science in which information such as published literatures, experiments, DNA, RNA, protein sequences and computational analyses are organized and stored. They are of important in allowing public and free access to biological data, sharing previous work by other scientists among others. Biological databases are categorized widely into;
-
Primary
-
Secondary
-
Composite databases
Primary databases
Contains mostly non curated DNA and RNA sequences from sequencing facilities.
Below is a list of common primary databases.
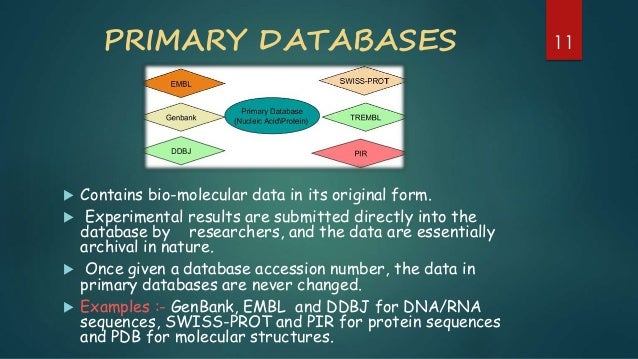
Secondary databases
They consist of curated DNA, RNA and protein sequences derived form primary databases.
Examples include Uniprote, pfam, PROSITE among others.
Composite databases
Consists of a combination of data derived from both primary and secondary databases.
Include OMIM, which consists mostly of human hereditary sequence data.
GENE, whose composition iS...