PBMC Example 1 - nolanlab/citrus GitHub Wiki
In the 2012 paper Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators, Bodenmiller et al. used mass cytometry to measure PBMC's from 8 different patients after 11 different stimulation conditions as well as an unstimulated "Reference" state. In this example, we use Citrus to automatically identify cell types that respond to one of those stimulations, BCR-crosslinking.
1) Data from the paper is available at cytobank.org. Download, "Reference" and "BCR-XL" samples for each of the 8 patients and save them to a local directory on your computer. They should be named:
- PBMC8_30min_patient1_BCR-XL.fcs
- PBMC8_30min_patient1_Reference.fcs
- PBMC8_30min_patient2_BCR-XL.fcs
- PBMC8_30min_patient2_Reference.fcs
- PBMC8_30min_patient3_BCR-XL.fcs
- PBMC8_30min_patient3_Reference.fcs
- PBMC8_30min_patient4_BCR-XL.fcs
- PBMC8_30min_patient4_Reference.fcs
- PBMC8_30min_patient5_BCR-XL.fcs
- PBMC8_30min_patient5_Reference.fcs
- PBMC8_30min_patient6_BCR-XL.fcs
- PBMC8_30min_patient6_Reference.fcs
- PBMC8_30min_patient7_BCR-XL.fcs
- PBMC8_30min_patient7_Reference.fcs
- PBMC8_30min_patient8_BCR-XL.fcs
- PBMC8_30min_patient8_Reference.fcs

2) Start the R GUI and launch Citrus interface
library("citrus")
citrus.launchUI()
3) Select single file from directory where experiment data resides.

4) In Sample Group Setup tab, change name of Group 1 to "Reference" and Group 2 to "BCR-XL".
Verify files are assigned correctly in summary panel on left.

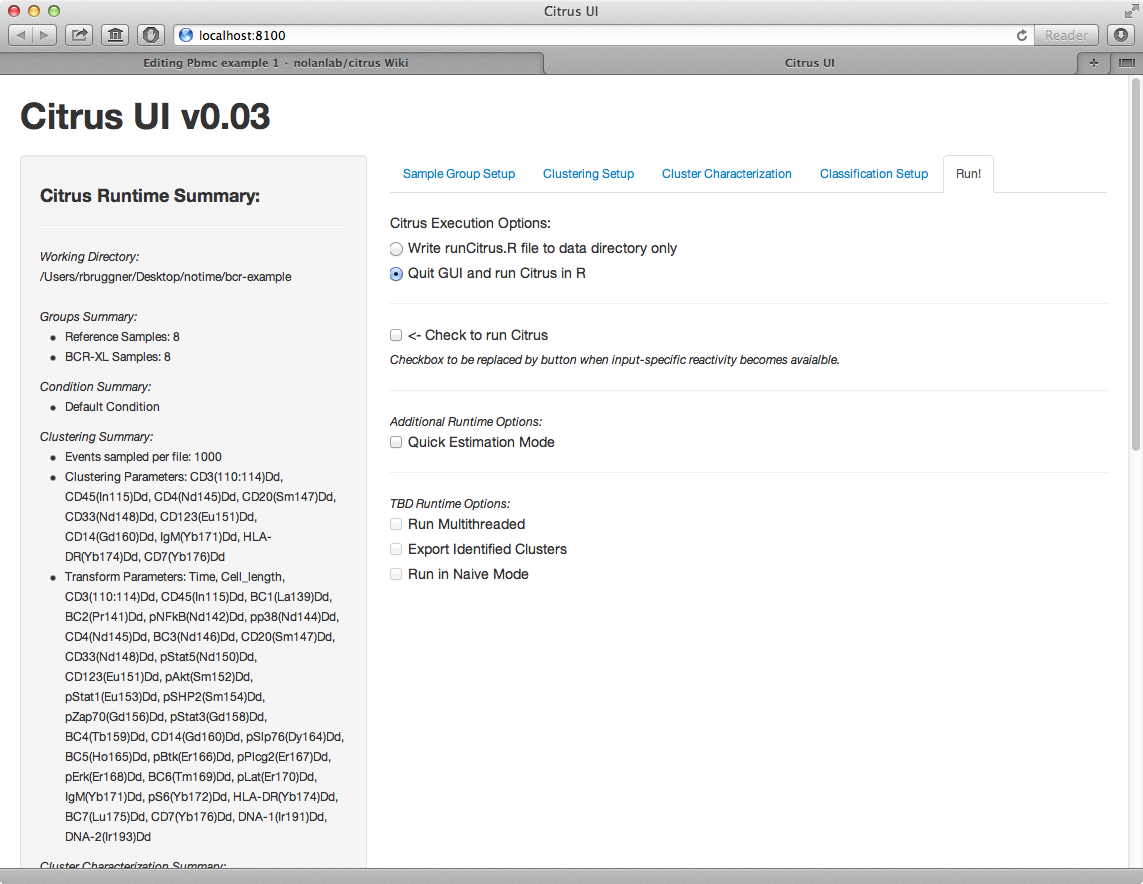
5) In clustering setup tab, set "Events sampled per file" to 5,000. For clustering parameters, select all CD markers, plus IgM and HLA-DR. For transform markers, select all markers except Time and Cell Length.

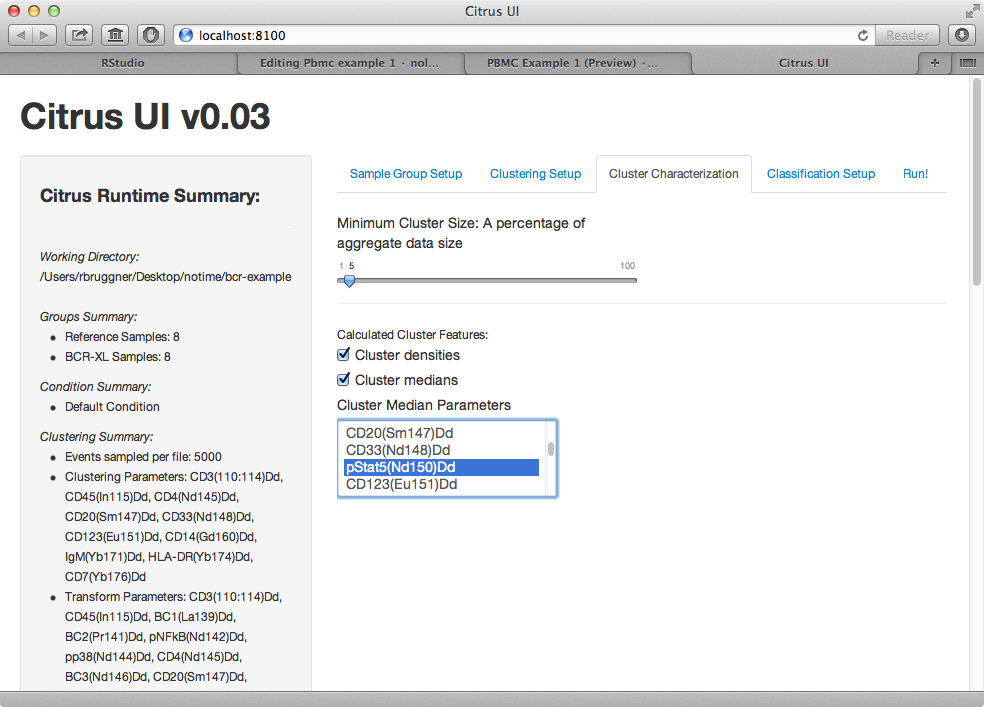
6) In clustering characterization tab, under Calculated Cluster Features, uncheck densities and check cluster medians. In the cluster median parameters select box, select all phosphoproteins (all markers starting with lowercase p).
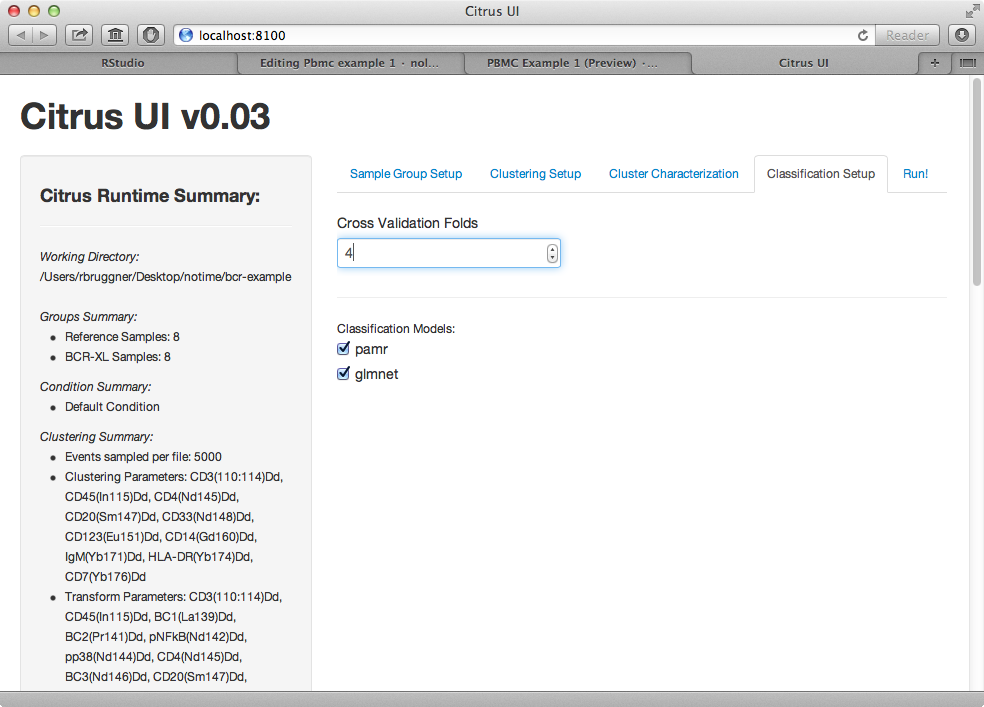
7) In classification setup tab, set cross-validation folds to 4.
8) In run tab, select "Quit UI and run Citrus in R" and check the "Run Citrus" checkbox.