Usage examples - labsquare/cutevariant GitHub Wiki
We will use the same protocol as SnpEff which is described on this page.
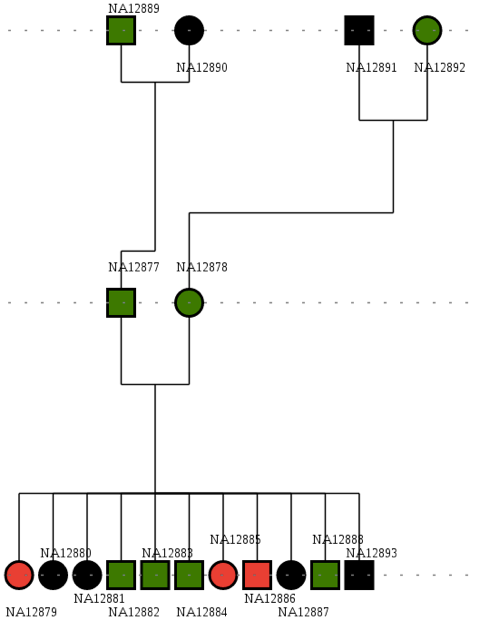
As mentioned, the aim is to find a pathogenic mutation in a family of 17 individus where 3 (in red) are affected by an homozygous inheritance disease.
Download dataset
You can download raw VCF file and annoate yourself using SnpEff or VEP.
But to make the example easier, we provide the SnpEff annotated file VCF available here.
Create a new project
After cutevariant is launched, click on new project on the upper toolbar and follow the wizard window instruction. depending of the size of the file, it can takes several minutes to be imported into sqlite database. But keep in mind, this step is performed only one time. Once the project has been created, you can open the sqlite file directly.

Select fields to display
You should now see the following view displaying a table of all variants loaded from the VCF file. Different plugins are available around to control the view and filtering in different way.
For instance, we selected here from the columns selector plugin, 7 differents fields (chr,pos,ref,alt,consequence,impact,gene).
You can choose as many fields as you wish. But displaying too much fields will be probably unreadable. Instead you can read fields value from of the current selected variant inside the InfoVariant plugin.

Filter variant with VQL
From the original SnpEff documentation, we have to find all variant with HIGH impact where three case are homozygous and the controls are not. Cutevariant generates special fields per variant telling the count of sample with a specific genotype. For instance the field "control_count_hom" return the number of sample cases who are homozygous. using those fields, you can then filters variants which are homozygous in the 3 samples from case group but not in the control group using the following VQL query.
SELECT favorite,comment,chr,ref,alt,consequence,impact,gene FROM variants
WHERE case_count_hom = 3.0 AND control_count_hom = 0.0 AND impact = 'HIGH'
We then find the variant in CFTR mentioned in the example of snpEff. You can then right-click on this variant to get more information about it.
