Home - jonathanbrecher/sharedclustering GitHub Wiki
Shared Clustering is a tool that allows an advanced or expert genetic genealogist to extract more information -- and more useful information -- from Ancestry DNA shared match lists. Rather than providing a single list of matches ordered only by the strength of the match, Shared Clustering divides that list into smaller clusters of matches that are likely related to each other. Knowing which matches are related to each other can often be a huge help in figuring out how those people are related to you, the test taker.
Click here to download the Shared Clustering application for Windows
If the previous link doesn't work for you, click here to download the same thing in *.zip format. But try the main link first. It seems to work fine for most people.
See also an introductory video, about 15 minutes long.
- Introduction
- Quick start for clustering
- The saved cluster diagram
- Tips for researching clusters
- Using the Shared Clustering application
- Interpreting clusters
- What is a cluster
- Clusters with red overlap
- Clusters with gray overlap
- Clusters with dark areas off of the diagonal
- Isolated clusters
- Close relatives
- What is a cluster
- Matches over 90 cM
- Matches over 50 cM
- Distant matches
- Endogamy
- Endogamy - Advanced analysis techniques
- Clusters represent shared segments
- Breaking brickwalls
- Caveats
- Frequently asked questions (FAQs)
- Administrivia
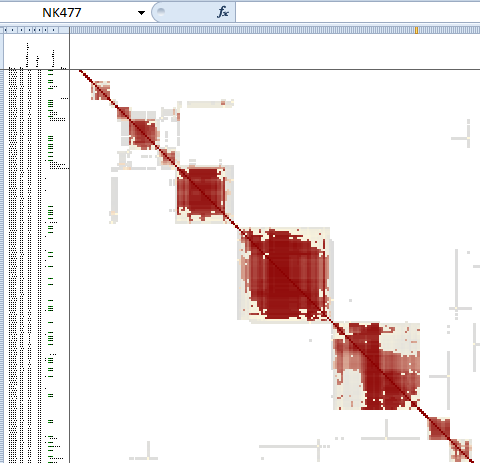
What is a cluster?
Each cluster contains a group of people who are more similar to each other than to other matches. Similarity is determined by the shared match lists of each match. In a perfect cluster, every member of the cluster appears in the shared match lists of every other member. Not all clusters are perfect. Clusters reflect biology, and biology is not perfect. The idea behind clusters is similarity, not perfection.
In most cases, a cluster represents a group of people who have some DNA segment shared among them. That makes sense. Clusters are built from the shared match lists reported by Ancestry, and Ancestry builds those lists from the DNA data.
Clusters are displayed visually as red squares in a spreadsheet, along with other identifying information about each match within the cluster: