merge_orthogroups - biologyguy/RD-MCL GitHub Wiki
This facilitates a semi-manual final polishing step, allowing you to test the appropriateness of folding a small orthogroup into a larger one when your knowledge of the protein family suggests RD-MCL may have broken up clusters erroneously. If you are convinced the clusters should be merged, this tool can also do that merge cleanly for you.
$: merge_orthogroups rdmcl_dir group_name <args>
rdmcl_dir: Path to an RD-MCL output directory.
group_name: Specify which orthogroup to test. Groups are named 'group_0_*'.
Always start by checking your query cluster. Running merge_orthogroups without the --merge flag will generate a report of how that cluster compares against all other larger clusters.
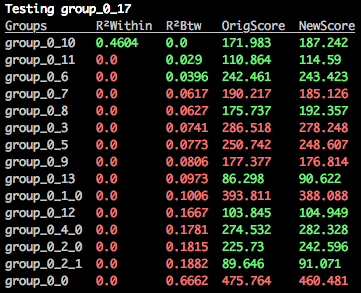
$: merge_orthogroups caspases group_0_17Two similarity metrics are used by RD-MCL. The first is based on a multiple sequence alignment and the relative similarity at each residue position (based on a substitution matrix and inferred secondary structure properties of the sequence). The second similarity metric is a correlation coefficient (R²) based on the output of the forward algorithm, as calculated for each sequence against hidden Markov models for each sequence. This matrix looks like the following snippet:
rec_id1 rec_id2 r_square
Psin-XP_006126818 Cstr-XP_010194245 0.02666175532
Psin-XP_006126818 Cmyd-XP_007054624 0.79362681219
Psin-XP_006126818 Tery-XP_009989658 0.02627408887
Psin-XP_006126818 Svul-XP_014734869 0.02116822948
Psin-XP_006126818 Tgut-XP_010222071 0.03274028803
Psin-XP_006126818 Cpug-XP_014817667 0.00365877299
These values are not used during Markov clustering, but invariably, sequences from the same cluster have high correlation coefficients (for example Psin-XP_006126818 and Cmyd-XP_007054624) while the values between clusters are low.
One continuous distribution is generated from all within-cluster scores and a second distribution is generated from the scores between the query cluster and the target cluster. The proportion of overlap between the two distributions is then assessed, and returned as the R²Within score. The closer to 1.0 the better, and anything below 0.05 is generally considered questionable.
Similar to R²Within, this value refers to the distribution overlap between the query/target cluster correlation scores and the scores found between all clusters. In this case, the closer to 0.0 the better, and anything above 0.05 should be considered questionable.
RD-MCL uses an optimality function to score the quality of a putative orthogroup. This is the sum score of the query and target cluster without merging.
This is the optimality score of the new cluster if the query and target are merged. If this value is lower than the original score, then it means you are probably combining paralogs. If you are certain that these are from recent gene duplications in a limited number of outlier taxa then it may make sense to merge, but be careful.
args: All flagged arguments are explained in detail below.
Provide the name of the group you want to merge into then follow the prompts. Merging will automatically update the final_clusters.txt file and other project data, as well as writing a log message to manual_merge.log
This will force a merge without any prompts. BE CAREFUL!!! Merging doesn't have an undo option.