homolog_tree_builder - biologyguy/RD-MCL GitHub Wiki
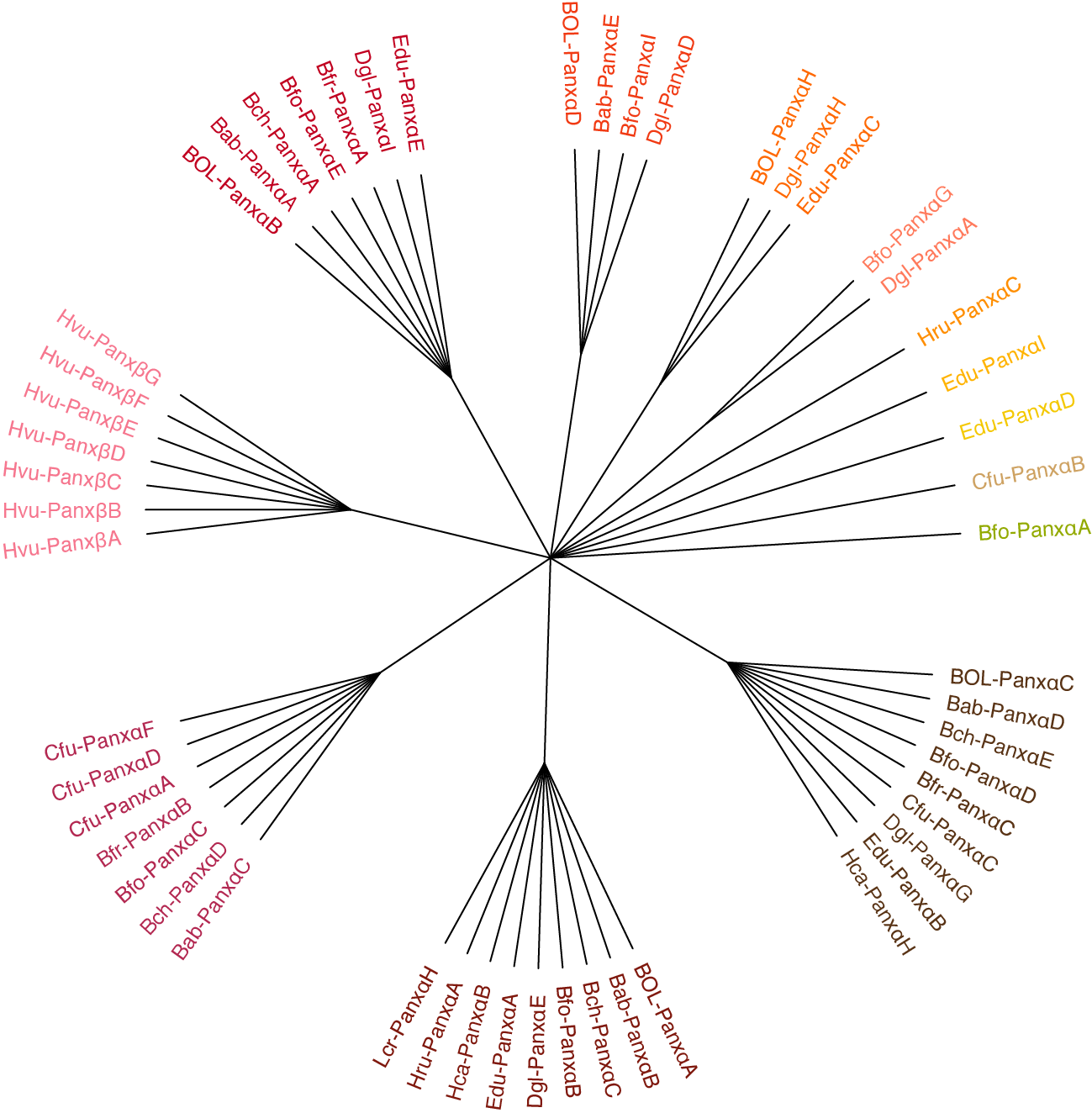
This tool Converts an RD-MCL final_clusters.txt file into a cladogram for better viewing. Each cluster
is arranged as if it were a polytomy, and if a cluster has been recursively disassociated, that structure
will be displayed in hierarchical fashion.
$: homolog_tree_builder cluster_file <args>
cluster_file: Each cluster is a whitespace-separated list of sequence IDs, one cluster per line, prefaced with
a group id and cluster score. This is the format output by rdmcl.py in `final_clusters.txt. For example:
group_0_1 70.0358 BOL-PanxαC Bab-PanxαD Bch-PanxαE Bfo-PanxαD Bfr-PanxαC Cfu-PanxαC Dgl-PanxαG Edu-PanxαB Hca-PanxαH
group_0_3 45.8238 BOL-PanxαA Bab-PanxαB Bch-PanxαC Bfo-PanxαB Dgl-PanxαE Edu-PanxαA Hca-PanxαB Hru-PanxαA Lcr-PanxαH
group_0_0 75.9846 Bab-PanxαC Bch-PanxαD Bfo-PanxαC Bfr-PanxαB Cfu-PanxαA Cfu-PanxαD Cfu-PanxαF
group_0_18 2.0734 Hvu-PanxβA Hvu-PanxβB Hvu-PanxβC Hvu-PanxβD Hvu-PanxβE Hvu-PanxβF Hvu-PanxβG
group_0_2 54.1235 BOL-PanxαB Bab-PanxαA Bch-PanxαA Bfo-PanxαE Bfr-PanxαA Dgl-PanxαI Edu-PanxαE
group_0_5 20.471 BOL-PanxαD Bab-PanxαE Bfo-PanxαI Dgl-PanxαD
group_0_6 13.037 BOL-PanxαH Dgl-PanxαH Edu-PanxαC
group_0_7 9.316 Bfo-PanxαG Dgl-PanxαA
group_0_19 2.9556 Hru-PanxαC
group_0_20 1.642 Edu-PanxαI
group_0_23 1.642 Edu-PanxαD
group_0_26 2.463 Cfu-PanxαB
group_0_30 1.4778 Bfo-PanxαA
args: All optional flagged arguments are explained in detail below.
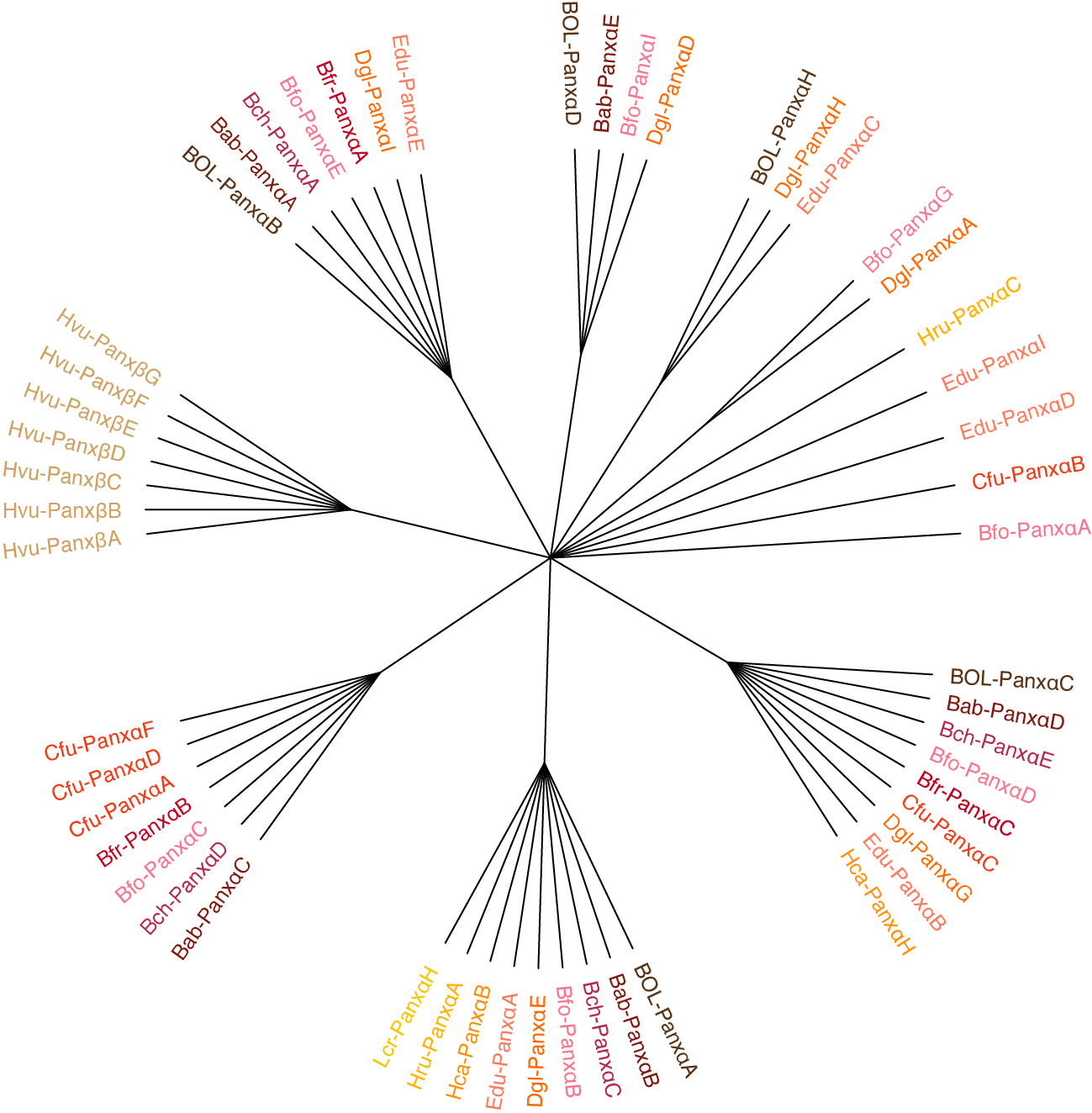
When this flag is set, the color of each branch tip will correspond to the species the sequence was derived from, not the cluster it is part of.
$: homolog_tree_builder '/path/to/final_clusters.txt' -cs
If you have used a non-standard separator to distinguish species from sequence ID, you can specify this here (default = '-').
$: homolog_tree_builder '/path/to/final_clusters.txt' -ts '~'