compare_homolog_groups - biologyguy/RD-MCL GitHub Wiki
This program is primarily for development purposes, allowing us to quickly visualize how well RD-MCL has performed on a well described data set.
$: compare_homolog_groups true_clusters_file rdmcl_clusters_file <args>
true_clusters_file: Path to curated clusters. Each cluster should be on a new line, and each sequence ID separated by whitespace.
$: cat true_clusters.tsv
BOL-PanxαE Bab-PanxαC Bch-PanxαD Bfo-PanxαC Bfr-PanxαB Cfu-PanxαA Cfu-PanxαB Cfu-PanxαD Cfu-PanxαF
BOL-PanxαC Bab-PanxαD Bch-PanxαE Bfo-PanxαD Bfr-PanxαC Cfu-PanxαC Dgl-PanxαG Edu-PanxαB Hca-PanxαH
BOL-PanxαA Bab-PanxαB Bch-PanxαC Bfo-PanxαB Dgl-PanxαE Edu-PanxαA Hca-PanxαB Hru-PanxαA Lcr-PanxαH
Hvu-PanxβA Hvu-PanxβB Hvu-PanxβC Hvu-PanxβD Hvu-PanxβE Hvu-PanxβF Hvu-PanxβG
BOL-PanxαB Bab-PanxαA Bch-PanxαA Bfo-PanxαE Bfr-PanxαA Dgl-PanxαI Edu-PanxαE
Bfo-PanxαA Bfo-PanxαG Dgl-PanxαA Edu-PanxαD Edu-PanxαI Hru-PanxαC
BOL-PanxαD Bab-PanxαE Bfo-PanxαI Dgl-PanxαD
BOL-PanxαH Dgl-PanxαH Edu-PanxαC
rdmcl_clusters_file: Path to RD-MCL final_clusters.txt file. These files can look a little bit different than the example above.
$: cat final_clusters.txt
group_0_1 70.0358 BOL-PanxαC Bab-PanxαD Bch-PanxαE Bfo-PanxαD Bfr-PanxαC Cfu-PanxαC Dgl-PanxαG Edu-PanxαB Hca-PanxαH
group_0_3 45.8238 BOL-PanxαA Bab-PanxαB Bch-PanxαC Bfo-PanxαB Dgl-PanxαE Edu-PanxαA Hca-PanxαB Hru-PanxαA Lcr-PanxαH
group_0_0 75.9846 Bab-PanxαC Bch-PanxαD Bfo-PanxαC Bfr-PanxαB Cfu-PanxαA Cfu-PanxαD Cfu-PanxαF
group_0_18 2.0734 Hvu-PanxβA Hvu-PanxβB Hvu-PanxβC Hvu-PanxβD Hvu-PanxβE Hvu-PanxβF Hvu-PanxβG
group_0_2 54.1235 BOL-PanxαB Bab-PanxαA Bch-PanxαA Bfo-PanxαE Bfr-PanxαA Dgl-PanxαI Edu-PanxαE
group_0_5 20.471 BOL-PanxαD Bab-PanxαE Bfo-PanxαI Dgl-PanxαD
group_0_6 13.037 BOL-PanxαH Dgl-PanxαH Edu-PanxαC
group_0_7 9.316 Bfo-PanxαG Dgl-PanxαA
group_0_19 2.9556 Hru-PanxαC
group_0_20 1.642 Edu-PanxαI
group_0_23 1.642 Edu-PanxαD
group_0_26 2.463 Cfu-PanxαB
group_0_30 1.4778 Bfo-PanxαA
args: All flagged arguments are explained in detail below.
Output a table with a set of similarity metrics.
| Metric | Description |
|---|---|
| Precision | TP / (TP + FP) |
| Recall | TP / (TP + FN) |
| Accuracy | ((TP + TN) / #seqs) * (#clusters / #seqs) |
| tn rate | (TN / (TN + FP)) * (#clusters / #seqs) |
| Query Score | Σ(RD-MCL cluster scores) |
| True Score | Σ(RD-MCL cluster scores) |
Where:
- TP = True positives
- FP = False positives
- TN = True negatives
- FN = False negatives
- #seqs = Total number of records included
- #clusters = Total number of clusters in the final_clusters file
$: compare_homolog_groups true_clusters_file rdmcl_clusters_file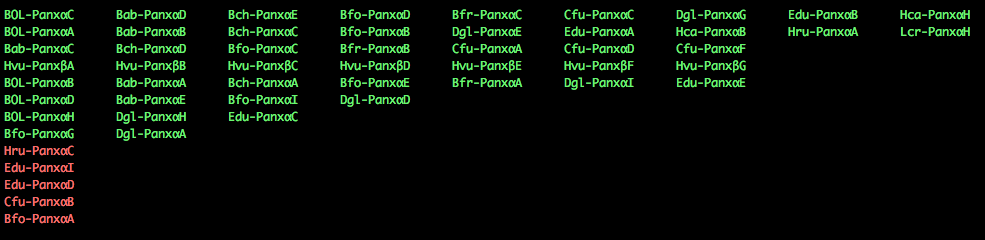
$: compare_homolog_groups true_clusters_file rdmcl_clusters_file --scorePrecision: 88.89%
Recall: 83.54%
Accuracy: 97.22%
tn rate: 97.98%
Query score: 121.21
True score: 119.94