PeptideShaker Online Frontend - barsnes-group/peptide-shaker-online GitHub Wiki
Introduction
PeptideShaker Online is a user-friendly web-based application with an intuitive easy-to-use interface. This makes it possible for users to process their data without the need for advanced programming skills or the use of command lines. The frontend supports a coordinated interactive visualization for mass spectrometry-based proteomics data, combined with dividing the data into three separate levels (Dataset Overview, Protein Overview, and Peptide-Spectrum Matches).
Frontend design
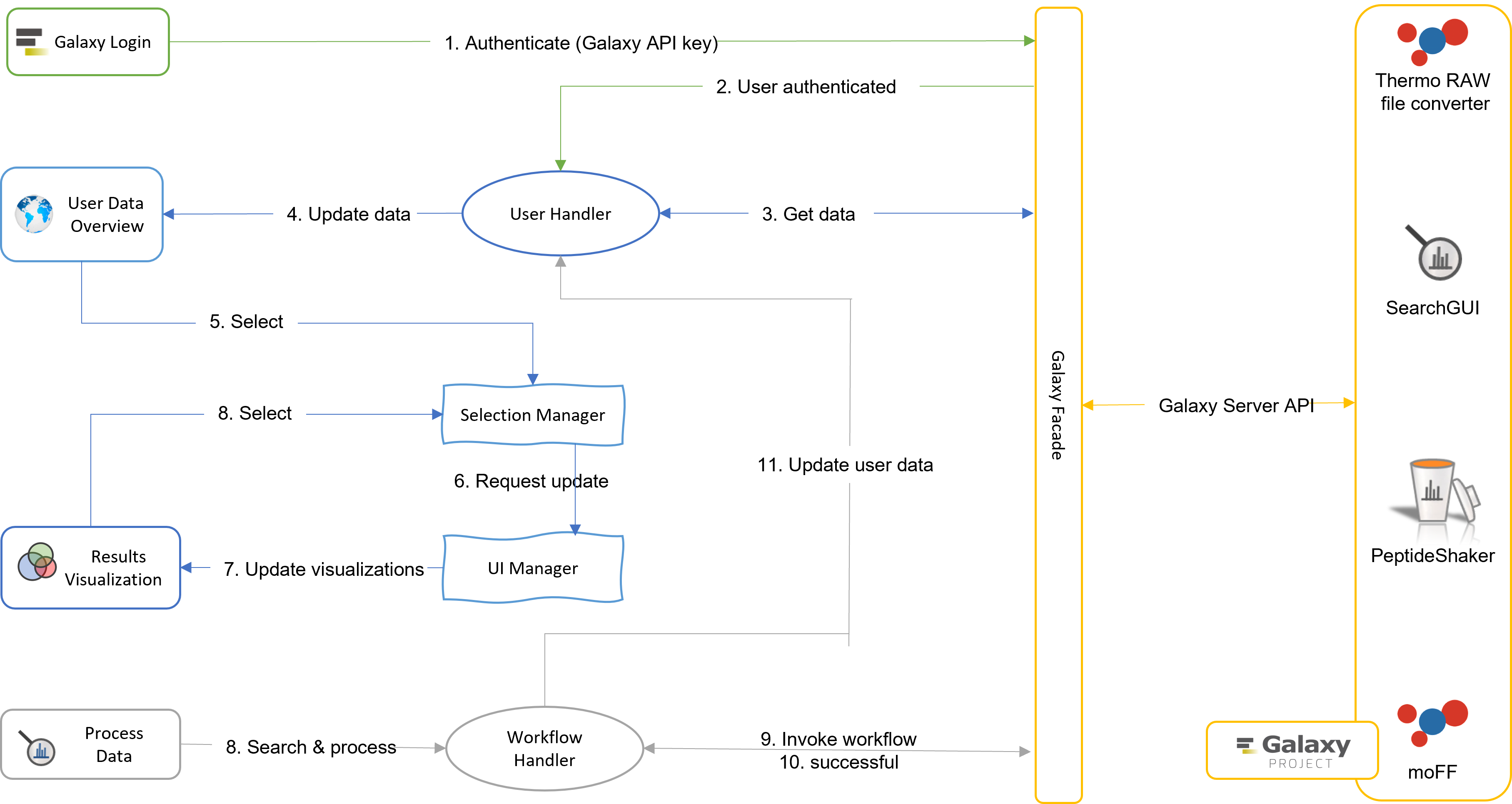
The frontend design includes five main classes: (i) GalaxyFacade.class - interacting with the Galaxy server platform using the Galaxy API service; (ii) UserHandler.class - user data handling including updating user datasets files; (iii) WorkflowHandler.class - data search and processing; (iv) SelectionManager.class - dataset/protein/peptide selections and filtering; and (v) UIManager.class - coordinating and updating the interactive visualizations.
Implementation and libraries
The frontend was developed using Vaadin 7.26 and Java 8. The demo is deployed on Tomcat server version 9 as a Java servlet container. The system uses compomics-utilities v5.0.15 to produce the main spectrum charts.
It also relies on the following libraries: biojava-core-5.4.0, commons-codec-1.10, javax.ws.rs-ap 2.1.1, javax.servlet-api 4.0.1, jersey (client,jersey-hk2,jersey-common,jersey-media-multipart)2.34, jersey-container-servlet-core 2.6, jettison 1.4.1, jfreesvg 3.3, jsclipboard 1.0.12, jung (graph-impl and visualization) 2.0.1, mysql-connector-java 8.0.16, plupload-vaadin 2.1.0, simplefiledownloader 1.0.4, sizereporter 0.1.4, tableexport-for-vaadin 1.6.2 and vertx-core 3.5.4.
See the source code for additional details.