GenomeMetrics - aechchiki/SIB_LongReadsWorkshop_Zurich18 GitHub Wiki
Genome metrics
Section: Genome assembly assessment [3/5].
Continuity
A very popular metrics used to evaluate assemblies is a wighted median of contig sizes, called N50. It represents a size of smallest contig in a set of contigs covering at least half of genome :
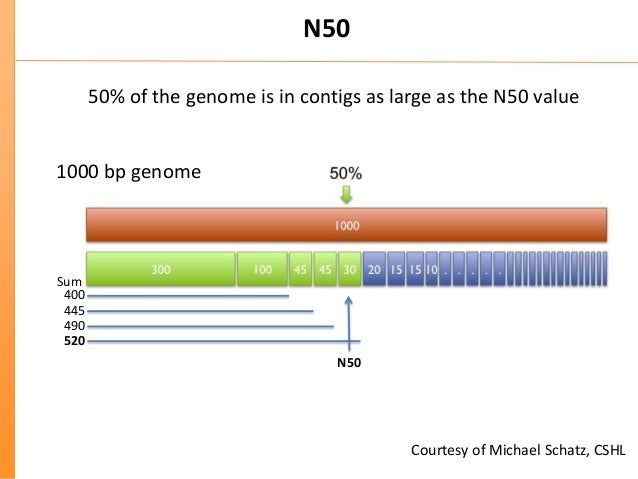
Calculation of N50
Let the set of assembled contigs be sorted from the longest to shortest contig. Now we sum contig sizes till we reach an half of total assembly size (i.e. sum of all contig sizes in the assembly). The last contig size that we added to summation is N50.
Note that this metrics rely on completely correct assembly. If all the reads would be just catenated in one huge (completely wrong) contig, N50 will be huge even the assembly is meaning less. N50 should never be the only metric used to evaluate assemblies.
NG50
A baby step towards biological reality is using known genome size instead of total sum of contigs for calculation of N50, such metrics is called then NG50.
Completeness
The idea here is to build approximate set of expected genes that can be found in assembly. The very first approach Cegma have identified a dataset of ultra conserved eukaryotic genes that was used as set of genes that should be identified in every Eukaryotic assembly. This software is no longer supported, it has been replaced by BUSCO. Developers of BUSCO provide set of conserved single-copy genes for several major taxonomic groups. The conserved genes in BUSCO are defined as genes that are shared in a unique copy at least by 90% of species in a taxa, which help sets to be rather big. However, at the same time it means that identification of all of them in a single copy might be simply impossible because of biological reality.
Note that genes are supposed to be in a single copy. Therefore extensive amount of genes identified as duplicated might be a good indication of partial of complete separation of haplotypes in the assembly.
Here an example of BUSCO output:
INFO Results:
INFO C:17.9%[S:17.8%,D:0.1%],F:0.1%,M:82.0%,n:1440
INFO 258 Complete BUSCOs (C)
INFO 257 Complete and single-copy BUSCOs (S)
INFO 1 Complete and duplicated BUSCOs (D)
INFO 1 Fragmented BUSCOs (F)
INFO 1181 Missing BUSCOs (M)
INFO 1440 Total BUSCO groups searched