Results File Structure - UVA-CAMA/NICUHDF5Viewer GitHub Wiki
An Overview of the Results File Structure
(HDF5 Viewer v3.1, BAP v1.2, 5/1/19)
The results file structure can be a bit confusing, but it is easy enough to explain.
The results file structure has a few different variables in it:
- info
- result_data
- result_name
- result_qrs
- result_tagcolumns
- result_tags
- result_tagtitle
The results file contains four different types of data, and each of these variables fits into one of these categories:
- Tag information
- result_tagcolumns
- result_tags
- result_tagtitle
- Continuous result data
- result_data
- result_name
- QRS detection information
- result_qrs
- Information about the original file
- info
This may seem obvious when I list it out like this, but it is helpful to understand which variables always go together.
Tag Information:
Let’s start with explaining how tag information works. For most of the algorithms we use (though not all), some sort of tag rule is included. Tags mark distinct events. Tags do not contain information about the continuous output from an algorithm. For the brady detection algorithm, we of course tag times where the HR is below a certain value. However, for the periodic breathing algorithm, we generate a continuous probability of periodic breathing and then tag the output from the algorithm with our binary tags, only marking events where the probability of periodic breathing is above 0.6.
result_tagtitle contains the names of all of the algorithms that have been run, with the exception of the QRS detection algorithms. The first column contains ‘/Results/Abbreviated name of algorithm.’ The second column contains the algorithm version number.
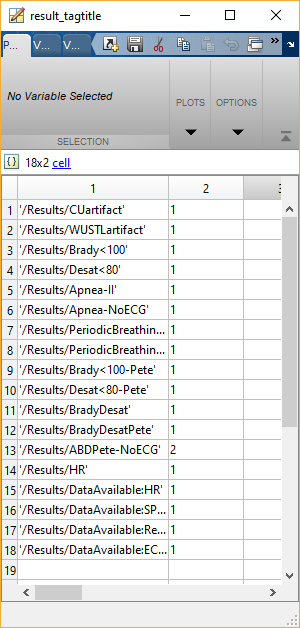
result_tags contains the actual tag information from all of the tagged events. The rows in result_tags correspond with the tag titles in result_tagtitle. result_tags(row).tagtable contains a table of the information about each individual event detected by that algorithm. This table has a number of rows equivalent to the number of detected events and may have a different number of columns depending on the algorithm. Some algorithms may yield an empty matrix here. That could be because no events were detected or because that algorithm does not return a tag (i.e. the HR “algorithm” simply stores a copy of the HR signal but does not look for any events, therefore this tagtable row will always have an empty set of brackets). In order to determine what each column means, we need to look at result_tagcolumns.

result_tagcolumns provides the labels for the columns in result_tags.tagtable. Again, the row within result_tagcolumns lines up with the row within result_tags and result_tagtitle. When you go to result_tagcolumns(row).tagname, you can see the names of the columns that correspond to the columns in result_tags(row).tagtable.

Continuous Result Data:
Only some algorithms output continuous result data. Algorithms like the periodic breathing algorithm outputs a continuous probability of periodic breathing at every time point (well, technically for periodic breathing, it doesn't return one every time point, but rather on a time interval, but that is a detail). Algorithms that output continuous data may or may not also output event tags, as we explained earlier regarding the periodic breathing algorithm which tags values >0.6. For algorithms which would yield only a binary continuous timeseries, such as bradycardia events, we no longer (as of BAP v1.1 and HDF5Viewer v3.0) store the binary timeseries (we used to in previous versions, but it made the results files too big). Instead, within the HDF5Viewer, we create the binary timeseries from the tags stored for these algorithms. This means that when you look for result_data for algorithms with binary output (like bradycardia events), the result_data structure will be empty.
result_name is very similar to result_tagtitle, and in many cases would be identical. The first column contains ‘/Results/Abbreviated name of algorithm.’ The second column contains the algorithm version number.

result_data has the same number of rows as result_name, as each row of result_data corresponds to the algorithm referenced in result_name. result_data has two columns: data and time. The time arrays are not necessarily identical for each signal. Much of result_data may contain empty matrices. This is, again, because we are no longer storing the continuous version of the binary tag output in this structure.
QRS Detection Information:
The results from the QRS detection algorithm are stored in a completely different way than both the continuous result data and the tag data.
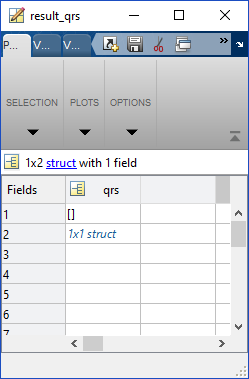
result_qrs can have a row for ECG Lead I, ECG Lead II, and ECG Lead III. The first row will always be for ECG I, the second for ECG II, and the third row for ECG III. It will only have as many rows as the largest ECG lead number it runs. Here is an example from a dataset that only contained ECG Lead II:
When you open result_qrs(lead).qrs, you will see the following structure:
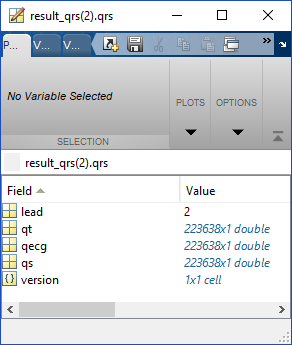
This structure gives you
- lead: the EKG lead number
- qt: the times of the detected beats
- qecg: the amplitude of the EKG signal at the time of the detected beat
- qs: this has to do with how Doug did the processing, and I don’t remember what it tells you, but it isn’t used in the algorithm
- version: the qrs detection algorithm version number
result_qrs is used in the HDF5Viewer to display the pink asterisks on top of the EKG signal when the EKG signal is plotted. It is also required for the versions of the apnea and periodic breathing algorithms which have an EKG lead number next to them
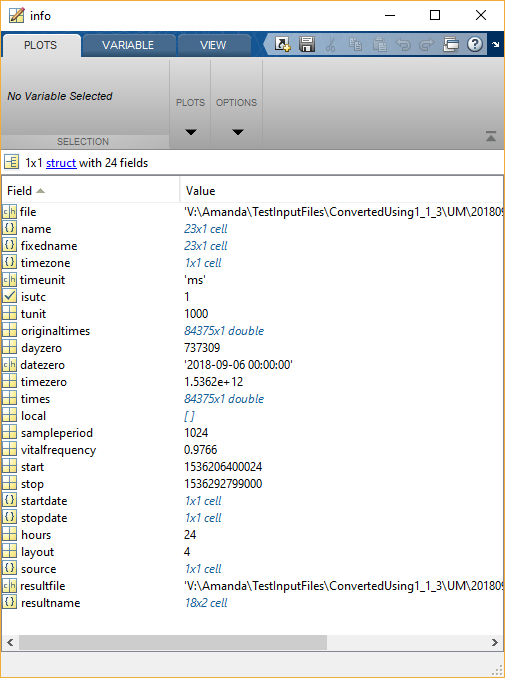
Information about the original file:
info contains a variety of information about the original file. This is handy to have around so that you don’t need to re-read the original file to find this information.