Fireball GPU - ProkopHapala/FireCore GitHub Wiki
Eigenvalue Problem
- Eigensolver - Jacobi-Rotation method can be efficiently paralelized:
Assembling:
ToDo:
- Check How many interpolations is called in which assemble functions.
- Notice that most of assembling is done just once per SCF-cycle
- Seems that
average_rho()is the most costly function
Ordering of arrays
Splines in interpolate_1d() are organized in such way that interaction and isub determines jxx which is 3d index after atomic types in1,in2 see splineint_2c(1,non2c,imid,jxx,in1,in2) in
subroutine interpolate_1d (interaction, isub, in1, in2, non2c, ioption, xin, yout, dfdx)
...
jxx = ind2c(interaction,isub)
...
aaa=splineint_2c(1,non2c,imid,jxx,in1,in2)
bbb=splineint_2c(2,non2c,imid,jxx,in1,in2)
ccc=splineint_2c(3,non2c,imid,jxx,in1,in2)
ddd=splineint_2c(4,non2c,imid,jxx,in1,in2)
Inside interpolate_1d_vec() we keep it, see
subroutine interpolate_1d_vec (interaction, isub, in1, in2, nME, ioption, xin, yout, dfdx)
aaa(:)=splineint_2c(1,:nME,imid,jxx,in1,in2)
bbb(:)=splineint_2c(2,:nME,imid,jxx,in1,in2)
ccc(:)=splineint_2c(3,:nME,imid,jxx,in1,in2)
ddd(:)=splineint_2c(4,:nME,imid,jxx,in1,in2)
In trescentros() we have it different, there are the ifs like
subroutine trescentros_vec (interaction, isorp, maxtype, in1, in2, indna, x, y, cost, eps, bcnax, nspecies)
if (interaction .eq. 1) then
hx = hx_bcna(isorp,index)
hy = hy_bcna(isorp,index)
nx = numx3c_bcna(isorp,index)
ny = numy3c_bcna(isorp,index)
xxmax = x3cmax_bcna(isorp,index)
yymax = y3cmax_bcna(isorp,index)
do iME = 1, index_max3c(in1,in2)
call interpolate_2d_vec (x, y, kforce, nx, ny, hx, hy, bcna_vec(:,1,1,iME,isorp,index), Q_L, dQ_Ldx, dQ_Ldy)
bcnalist(:,iME) = Q_L(:)
end do
else if (interaction .eq. 2) then
Questions
- Is somewhere actually used full 3-ceter
in1,in2,in3?
- Answer: - this is actually done by
common_neighbors(), we loop already over pairs of common neighbors like this:
do ialp = iatomstart, iatomstart - 1 + natomsp
rna(:) = ratom(:,ialp)
indna = imass(ialp)
do ineigh = 1, neigh_comn(ialp)
mneigh = neigh_comm(ineigh,ialp)
iatom = neigh_comj(1,ineigh,ialp)
jatom = neigh_comj(2,ineigh,ialp)
r1(:) = ratom(:,iatom) + xl(:,ibeta)
r2(:) = ratom(:,jatom) + xl(:,jbeta)
- What value have
index_max2c(in1,in2),index_max3c(in1,in2),index_maxPP(in1,in2),index_maxS(in1,in2)?- used e.g. in
doscentros(),doscentrosPP(),doscentrosS()like
- used e.g. in
do index = 1, index_max2c(in1,in3)
call interpolate_1d (interaction, isub, in1, in2, index, iforce, distance, slist(index), dslist(index))
end do
- used e.g. in
trescentros(),trescentrosS(),Dtrescentros(),DtrescentrosS()`
do iME = 1, index_maxS(in1,in2)
call interpolate_2d (x, y, kforce, nx, ny, hx, hy, den3S_01(1,1,iME,isorp,index), Q_L, dQ_Ldx, dQ_Ldy)
call interpolate_2d (x, y, kforce, nx, ny, hx, hy, den3S_02(1,1,iME,isorp,index), Q_L, dQ_Ldx, dQ_Ldy)
call interpolate_2d (x, y, kforce, nx, ny, hx, hy, den3S_03(1,1,iME,isorp,index), Q_L, dQ_Ldx, dQ_Ldy)
call interpolate_2d (x, y, kforce, nx, ny, hx, hy, den3S_04(1,1,iME,isorp,index), Q_L, dQ_Ldx, dQ_Ldy)
call interpolate_2d (x, y, kforce, nx, ny, hx, hy, den3S_05(1,1,iME,isorp,index), Q_L, dQ_Ldx, dQ_Ldy)
end do
Answer:
index_max2c
H-H 1
C-H 2
C-C 6
index_max3c
H-H 1
C-H 3
C-C 10
index_maxPP
H-H 1
C-H 2
C-C 6
index_maxS
H-H 1
C-H 2
C-C 4
- What are the value of
hx = hx_bcna(isorp,index)
hy = hy_bcna(isorp,index)
nx = numx3c_bcna(isorp,index)
ny = numy3c_bcna(isorp,index)
xxmax = x3cmax_bcna(isorp,index)
yymax = y3cmax_bcna(isorp,index)
used for interpolate_2d () ? Do they differ with different atoms and different interactions ?
Answer: Actually for all combinations of species and interactions are the same
hx = 0.17009285714285713
hy = 0.17009285714285713
nx = 29
ny = 29
xxmax = 4.7625999999999999
yymax = 4.7625999999999999
Overall structure of assemble_mcweda()
! ============================= FROM assemble_mcweda ()
if ((itheory .eq. 1 .or. itheory .eq. 2) .and. Kscf .eq. 1) then
call get_ewald ! (nprocs, my_proc, kforce, icluster, itheory, iordern)
end if
if(itheory_xc .eq. 2 ) then
call assemble_olsxc_1c ! no-interpolation
endif
if (Kscf .eq. 1) then
call assemble_sVNL() ! doscentrosPP()
call assemble_2c() ! doscentros()
call assemble_2c_PP() ! no-interpolation
end if
if ( (itheory_xc .eq. 1) .or. (itheory_xc .eq. 2) ) then
!write (*,*) ' Assemble SN-xc exchange-correlation interactions. '
if (itheory .eq. 1) then
call average_ca_rho() ! doscentros(), doscentrosS() ... really complicated function
else
call average_rho() ! doscentros(), doscentrosS() ... really complicated function
endif
end if
if (itheory_xc .eq. 1) then
call assemble_snxc_on () ! no-interpolation
call assemble_snxc_off() ! no-interpolation
else if (itheory_xc .eq. 2 ) then
call assemble_olsxc_on() ! no-interpolation
call assemble_olsxc_off() ! doscentros()
end if ! if (itheory_xc = 2)
if (itheory .eq. 1) then
call assemble_ca_2c() ! doscentros()
endif
if (Kscf .eq. 1) then
call assemble_3c() ! trescentros()
call assemble_3c_PP() ! no-interpolation
end if
if (itheory .eq. 1) then
call assemble_ca_3c() ! trescentros()
call assemble_lr () ! no-interpolation
else
Put all loop together
All assemble loops looks like this:
do iatom = iatomstart, iatomstart - 1 + natomsp
do ineigh = 1, neighn(iatom)
! do something like:
A1) call doscentros (interaction, isorp, kforce, in1, in2, in3, y, eps, deps, sx, spx)
A2) call interpolate_1d (interaction, ideriv, in1, in2, index, iforce, distance, slist(index), dslist(index))
A3) call doscentrosPP (interaction, isorp, y, eps, deps, iforce, in1, in2, sVNLx, spVNLx)
A4) call trescentros (interaction, isorp, isorpmax, in1, in2, indna, x, y, cost, eps, bcnax, nspecies)
B1) s_mat(imu,inu,ineigh,iatom) = sx(imu,inu)
B2) vnl(imu,inu,kneigh,iatom) = vnl(imu,inu,kneigh,iatom) + PPx(imu,inu)
B3) PPx(imu,inu) = PPx(imu,inu) + cl(ncc)*sVNL(imu,ncc,ineigh,iatom) *sVNL(inu,ncc,mneigh_self,jatom)
B4) bcnlx(imu,inu) = bcnlx(imu,inu) + cl(ncc)*sVNL(imu,ncc,m31,iatom)*sVNL(inu,ncc,m32,jatom)
B5) vna(imu,inu,mneigh,iatom) = vna(imu,inu,mneigh,iatom) + bcnax(imu,inu)*eq2
B6) sub_ewald(iatom) = sub_ewald(iatom) + dQ*ewald(iatom,jatom)
B7) ewaldlr(imu,inu,ineigh,iatom)=ewaldlr(imu,inu,ineigh,iatom)+(sterm-dterm)*sub_ewald(iatom)*eq2+ (sterm+dterm)*sub_ewald(jatom)*eq2
B8) h_mat(imu,inu,ineigh,iatom)=t_mat(imu,inu,ineigh,iatom)+vna(imu,inu,ineigh,iatom)+vxc(imu,inu,ineigh,iatom)+ vca(imu,inu,ineigh,iatom)
end do
end do
2-center terms:
Question: what value have index_max2c(in1,in3)
assemble_usr()
interpolate_1d()
doscentros()
do index = 1, index_max2c(in1,in3)
call interpolate_1d (interaction, isub, in1, in2, index, iforce, distance, slist(index), dslist(index))
end do
doscentrosPP()
do index = 1, index_maxPP(in1,in2)
call interpolate_1d (interaction, isub, in1, in2, index, iforce, distance, pplist(index), dpplist(index))
end do
doscentrosS_()
do index = 1, index_maxS(in1,in3)
call interpolate_1d (interaction, isub, in1, in2, index, iforce, distance, slist(index), dslist(index))
end do
3-center terms:
NOTE 1: we can significantly vectorize 3-center interactions by calling, there is the loop
do iME = 1, index_maxS(in1,in2)
call interpolate_2d (x, y, kforce, nx, ny, hx, hy, den3S_01(1,1,iME,isorp,index), Q_L, dQ_Ldx, dQ_Ldy)
call interpolate_2d (x, y, kforce, nx, ny, hx, hy, den3S_02(1,1,iME,isorp,index), Q_L, dQ_Ldx, dQ_Ldy)
call interpolate_2d (x, y, kforce, nx, ny, hx, hy, den3S_03(1,1,iME,isorp,index), Q_L, dQ_Ldx, dQ_Ldy)
call interpolate_2d (x, y, kforce, nx, ny, hx, hy, den3S_04(1,1,iME,isorp,index), Q_L, dQ_Ldx, dQ_Ldy)
call interpolate_2d (x, y, kforce, nx, ny, hx, hy, den3S_05(1,1,iME,isorp,index), Q_L, dQ_Ldx, dQ_Ldy)
end do
NOTE 2: We can merge trescentrosS() and trescentros()
NOTE 3: rotate_fb() and twister() is worth to optimize
rotate_fb()
call twister (eps, dmat, pmat)
do issh = 1, nssh(in1)
call chooser (l1, dmat, pmat, left)
do jssh = 1, nssh(in2)
l2 = lssh(jssh,in2)
call chooser (l2, dmat, pmat, right)
do m2 = 1, 2*l2 + 1
do m1 = 1, 2*l1 + 1
do k2 = 1, 2*l2 + 1
do k1 = 1, 2*l1 + 1
xmatrix(n1+k1,n2+k2) = xmatrix(n1+k1,n2+k2) + left(k1,m1)*mmatrix(n1+m1,n2+m2)*right(k2,m2)
end do
end do
end do
end do
end do
end do
twister()
do imu = 1, 5
xlambda11 = 0
xlambda12 = 0
xlambda13 = 0
xlambda32 = 0
xlambda33 = 0
do jx = 1, 3
do ix = 1, 3
if (amat(ix,jx,imu) .ne. 0.0d0) then
amat_term = amat(ix,jx,imu)
xlambda11 = xlambda11 + amat_term*eps(jx,1)*eps(ix,1)
xlambda12 = xlambda12 + amat_term*eps(jx,1)*eps(ix,2)
xlambda13 = xlambda13 + amat_term*eps(jx,1)*eps(ix,3)
xlambda32 = xlambda32 + amat_term*eps(jx,3)*eps(ix,2)
xlambda33 = xlambda33 + amat_term*eps(jx,3)*eps(ix,3)
end if
end do
end do
! Set the D matrices:
dmat(imu,1) = 2.0d0*xlambda12
dmat(imu,2) = 2.0d0*xlambda32
dmat(imu,3) = sqrt(3.0d0)*xlambda33
dmat(imu,4) = 2.0d0*xlambda13
dmat(imu,5) = 2.0d0*xlambda11 + xlambda33
end do
assemble_3c()
do ialp = iatomstart, iatomstart - 1 + natomsp ! loop over atoms in central cell
do ineigh = 1, neigh_comn(ialp) ! loop over neighbors
epsilon(R1,R2,spe) ! metric tens
trescentros() ! interpolation of integrals
trescentrosS_vec() ! interpolation of integrals
if(ivec_3c == 1)
do iME = 1, index_max3c(in1,in2)
interpolate_2d_vec()
end do
if(ivec_3c == 2)
t_data3c_interpolate_2d()
else
do iME = 1, index_max3c(in1,in2)
interpolate_2d() [ 5 x 3 ]
end do
Dtrescentros()
Dtrescentros_vec
do iME = 1, index_max3c(in1,in2)
interpolate_2d()
end do
DtrescentrosS()
DtrescentrosS_vec()
do iME = 1, index_maxS(in1,in2)
interpolate_2d()
end do
assemble_3c_PP()
NOTES : Indexes of interactions
From READFILES/read_2c.f90, used in interpolate_1d()
1 overlap
2 vna_ontopl
3 vna_ontopr
4 vna_atom
5 vnl
6 xc_ontop
7 xc_atom
8 xc_corr
9 dipole_z
10 dipole_y
11 dipole_x
12 coulomb
13 kinetic
14 nuxc
15 den_ontopl
16 den_ontopr
17 den_atom
18 dnuxc_ol
19 dnuxc_or
20 denS_ontopl
21 denS_ontopr
22 denS_atom
23 overlapS
From READFILES/read_3c.f90, used in interpolate_2d()
1 bcna_01
2 xc3c_02
3 den3_01
4 den3S_01
Tests
Time spend in function
1 assemble_mcweda 43.68
2 solveH 1.32
3 denmat 4.02
4 getenergy_mcweda 0.06
11 getforces_mcweda 33.82
1 assemble_mcweda 43.68
5 assemble_mcweda_neighbors 0.79
7 assemble_mcweda_1c 0.03
8 assemble_mcweda_2C 29.32
9 assemble_mcweda_3C 13.32
6 get_ewald 0.02
10 buildH 0.18
8 assemble_mcweda_2C 29.32
20 assemle_2c(Kscf=1) 2.15
21 average_rho 21.11
22 assemble_olsxc_on 0.07
23 assemble_olsxc_off 2.68
24 assemble_ca_2c 3.30
9 assemble_mcweda_3C 13.32
31 assemble_3c 2.27
32 assemble_3c_PP 0.23
33 assemble_ca_3c 10.72
34 assemble_lr 0.09
11 getforces_mcweda 33.82
50 assemble_F 0.03
51 Dassemble_2c 0.89
52 Dassemble_2c_PP 0.26
53 Dassemble_ca_olsxc_on 0.19
54 Dassemble_ca_olsxc_2c 1.69
55 Dassemble_ca_2c 1.44
56 Dassemble_3c 5.36
57 Dassemble_3c_PP 1.54
58 Dassemble_ca_3c 9.81
59 Dassemble_lr 1.19
60 Dassemble_ca_olsxc_3c 11.41
Number of function calls

ncall_count
doscentros 2 062 488
doscentrosS 540 056
trescentros 1 998 511
trescentrosS 899 564
Dtrescentros 826 543
DtrescentrosS 313 580
epsilon 2 666 784
deps2cent 724 368
interpolate_1d 8 398 760
twister 7 849 772
twisterd 2 941 702
makeDmat 2 921 174
rotate_fb 4 887 542
assemble_sVNL 100
assemble_2c 100
assemble_2c_PP 100
average_ca_rho 288
average_rho 0
assemble_snxc_on 0
assemble_snxc_off 0
assemble_olsxc_on 288
assemble_olsxc_off 288
assemble_ca_2c 288
assemble_3c 100
assemble_3c_PP 100
assemble_ca_3c 288
assemble_lr 288
interpolate_1d 8 398 760
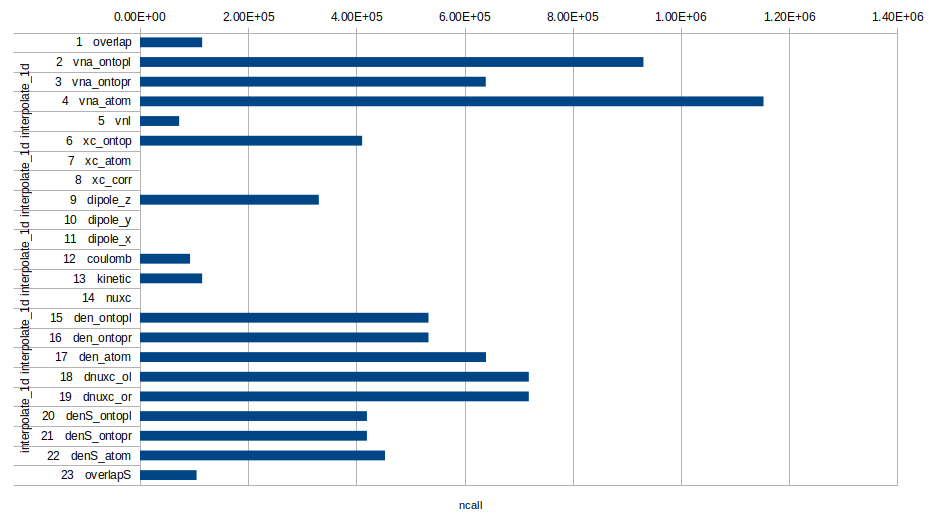
interpolate_1d 1 overlap 114 944
interpolate_1d 2 vna_ontopl 930 660
interpolate_1d 3 vna_ontopr 639 264
interpolate_1d 4 vna_atom 1 152 564
interpolate_1d 5 vnl 72 384
interpolate_1d 6 xc_ontop 410 600
interpolate_1d 7 xc_atom 0
interpolate_1d 8 xc_corr 0
interpolate_1d 9 dipole_z 330 576
interpolate_1d 10 dipole_y 0
interpolate_1d 11 dipole_x 0
interpolate_1d 12 coulomb 92 576
interpolate_1d 13 kinetic 114 944
interpolate_1d 14 nuxc 0
interpolate_1d 15 den_ontopl 533 320
interpolate_1d 16 den_ontopr 533 320
interpolate_1d 17 den_atom 639 608
interpolate_1d 18 dnuxc_ol 718 772
interpolate_1d 19 dnuxc_or 718 772
interpolate_1d 20 denS_ontopl 419 480
interpolate_1d 21 denS_ontopr 419 480
interpolate_1d 22 denS_atom 452 888
interpolate_1d 23 overlapS 104 608
interpolate_2d 79 468 260
interpolate_2d 1 bcna 37 280 810
interpolate_2d 2 xc3c 0
interpolate_2d 3 den3 27 763 600
interpolate_2d 4 den3S 14 423 850