1.2 Cache Design - PRIDE-Archive/ms-data-core-api GitHub Wiki
The general idea of caching is to reuse content to avoid repeating expensive operations. In the ms-data-core-api, this is a clear requirement since the average sizes of the experimental output files in proteomics experiments keep increasing. Therefore, loading the entire file in memory is no longer feasible in many cases. On the other hand, requesting the data content directly from the source file is often restricted by the file’s storage media. The processing cost for selecting a value from a file (e.g. spectrum, peptide or protein) is fairly high when compared to the cost of having the value stored in memory. So it seems plausible to use some caching mechanism that keeps frequently used values in the application instead of retrieving these values from the storage media every time. Most frameworks and tools have integrated caching mechanisms nowadays. One needs to balance between memory consumption and performance. A critical factor when using caching in Java is the size of the cache: when the cache grows too large, the Java Garbage Collector has to clean-up more often (which consumes time). This can lead to a gradual degradation of the performance, or the application may even crash if it exceeds the memory limit.
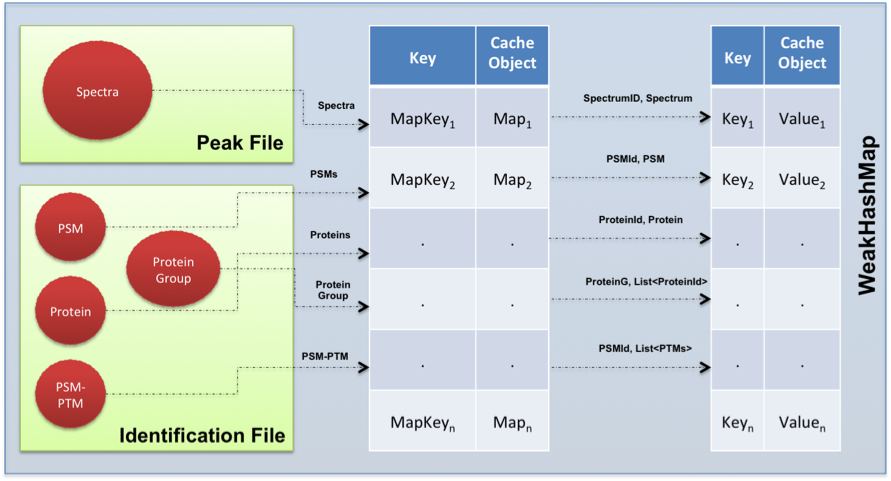
The ms-data-core-api controls the balance between the memory consumption and fast access to the data using two-level HashMaps. Most of the objects such as spectrum, peptide, and protein can be kept in memory for fast access. For this goal, most of the data structures in the API have a key-value representation. A global map is then used to group all the cache structures. The memory defines a cache size for each structure in order to avoid out-of-memory problems. Some values such as precursor ion mass and precursor ion charge are also stored in the cache since these values can frequently be accessed by third-party tools.
In the cache maps, not only complete objects are stored, the relationships between some frequently accessed data structures and their properties are also saved. This is one key feature of the library: it allows fast access to the data without the need to load the complete data structure in memory. For example, the cache modification map ensures fast access to the modifications of each PSM without the need to retrieve all the PSMs from the identification file.