1.5 Central dogma of molecular biology - Oronda/Molecular_biology_concepts GitHub Wiki
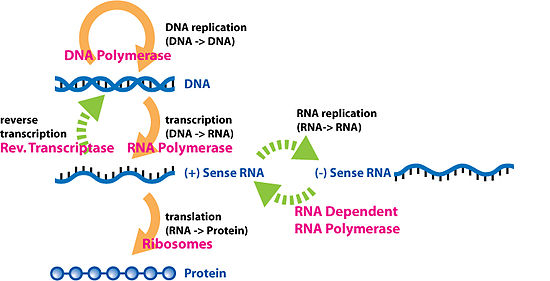
The 'central dogma' of molecular biology states that sequence information can be transferred among nucleic acids, and from nucleic acids to proteins, but sequence information cannot be transferred among proteins, or from proteins to nucleic acids. It states that DNA contains instructions for making a protein, which are copied by RNA. RNA then uses the instructions to make a protein. In short: DNA → RNA → Protein, or DNA to RNA to Protein
A.DNA Replication
The growth, development, and reproduction of organisms rely on cell division, or the process by which a single cell divides into two usually identical daughter cells. This requires first making a duplicate copy of every gene in the genome in a process called DNA replication. The copies are made by specialized enzymes known as DNA polymerases.
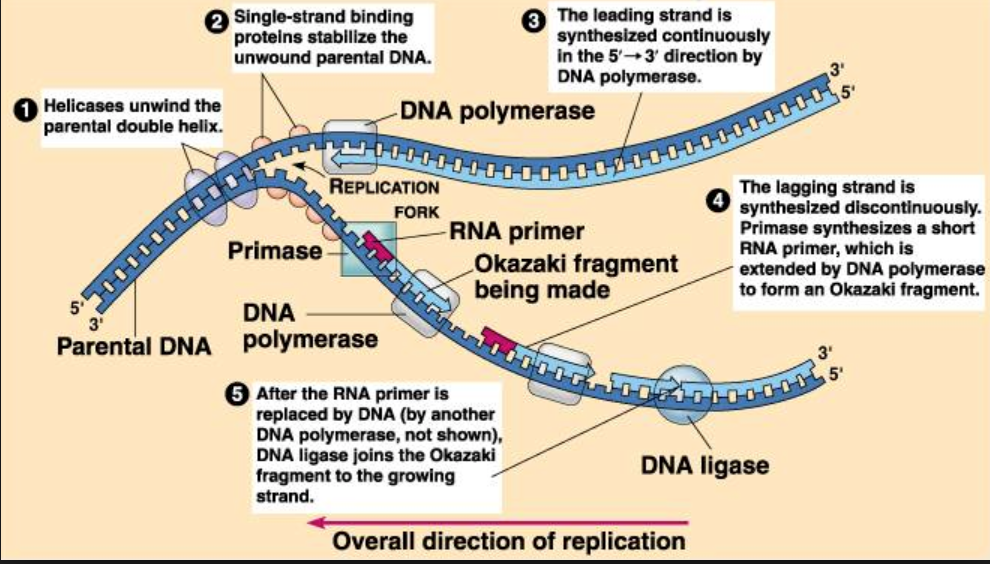
DNA replication begins with the "unzipping" of the parent molecule as the hydrogen bonds between the base pairs are broken. Once exposed, the sequence of bases on each of the separated strands serves as a template to guide the insertion of a complementary set of bases on the strand being synthesized.
The new strands are assembled from deoxynucleoside triphosphates. Each incoming nucleotide is covalently linked to the "free" 3' carbon atom on the pentose. The second and third phosphates are removed together as a molecule of pyrophosphate (PPi). The nucleotides are assembled in the order that complements the order of bases on the strand serving as the template. Thus, each C on the template guides the insertion of a G on the new strand, each G a C, and so on.
When the process is complete, two DNA molecules have been formed identical to each other and to the parent molecule.
Enzymes in DNA replication
- A portion of the double helix is unwound by a helicase.
- A molecule of a DNA polymerase binds to one strand of the DNA and begins moving along it in the 3' to 5' direction, using it as a template for assembling a leading strand of nucleotides and reforming a double helix. In eukaryotes, this molecule is called DNA polymerase delta (δ).
- Because DNA synthesis can only occur 5' to 3', a molecule of a second type of DNA polymerase (epsilon, ε, in eukaryotes) binds to the other template strand as the double helix opens. This molecule must synthesize discontinuous segments of polynucleotides (called Okazaki fragments).
- Another enzyme, DNA ligase I then stitches these together into the lagging strand.
B.Transcription
DNA transcription is the process by which the genetic information contained within DNA is re-written into messenger RNA (mRNA) by RNA polymerase. Transcription occurs when there is a need for a particular gene product at a specific time or in a specific tissue.
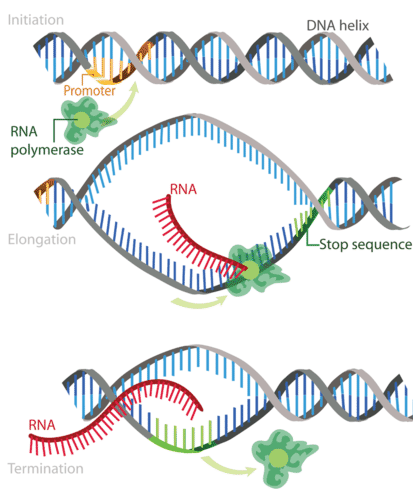
The process of DNA transcription can be split into 3 main stages: initiation, elongation & termination.
Initiation
Transcription is catalysed by the enzyme RNA polymerase, which attaches to and moves along the DNA molecule until it recognises a promoter sequence. This area of DNA indicates the starting point of transcription, and there may be multiple promoter sequences within a DNA molecule. Transcription factors are proteins that control the rate of transcription; they too bind to the promoter sequences with RNA polymerase. Once bound to the promoter sequence, RNA polymerase unwinds a portion of the DNA double helix, exposing the bases on each of the two DNA strands.
Elongation
One DNA strand (the template strand) is read in a 3′ to 5′ direction, and so provides the template for the new mRNA molecule. The other DNA strand is referred to as the coding strand. This is because its base sequence is identical to the synthesised mRNA, except for the replacement of thiamine bases with uracil. RNA polymerase uses incoming ribonucleotides to form the new mRNA strand. It does this by catalysing the formation of phosphodiester bonds between adjacent ribonucleotides, using complementary base pairing (A to U, T to A, C to G and G to C). Bases can only be added to the 3′ (three-prime) end, so the strand elongates in a 5’ to 3’ direction.
Termination
Elongation continues until the RNA polymerase encounters a stop sequence. At this point, transcription stops and the RNA polymerase releases the DNA template.
The mRNA which has been transcribed up to this point is referred to as pre-mRNA. Processing must occur to convert this into mature mRNA. This includes:
- 5′ Capping- Capping describes the addition of a methylated guanine cap to the 5′ end of mRNA. Its presence is vital for the recognition of the molecule by ribosomes, and to protect the immature molecule from degredation by RNAases.
- Polyadenylation- Describes the addition of a poly(A) tail to the 3′ end of mRNA. The poly(A) tail consists of multiple molecules of adenosine monophosphate. This stabilises RNA, which is necessary as RNA is much more unstable than DNA.
- Splicing-allows the genetic sequence of a single pre-MRNA to code for many different proteins, conserving genetic material. This process is sequence dependent and occurs within the transcript. It involves removal of introns (non-coding sequences) via spliceosome excision and joining together of exons (coding sequence) by ligation.
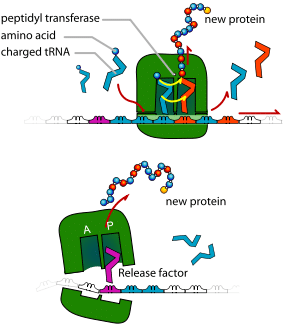
C.Translation
Translation is the process by which the genetic code contained within a messenger RNA (mRNA) molecule is decoded to produce a specific sequence of amino acids in a polypeptide chain.
The key components required for translation are mRNA, ribosomes, and transfer RNA (tRNA).
During translation, mRNA nucleotide bases are read as codons of three bases. Each codon codes for a particular amino acid. Every tRNA molecule possesses an anticodon that is complementary to the mRNA codon, and at the opposite end lies the attached amino acid. tRNA molecules are therefore responsible for bringing amino acids to the ribosome in the correct order, ready for polypeptide assembly.
A single amino acid may be coded for by more than one codon. There are also specific codons that signal the start and the end of translation.
Aminoacyl-tRNA synthetases are enzymes that link amino acids to their corresponding tRNA molecules. The resulting complex is charged and is referred to as an aminoacyl-tRNA.
Translation occurs in the cytoplasm following DNA transcription and, like transcription, has three stages: initiation, elongation and termination.
Initiation
For translation to begin, the start codon (5’AUG) must be recognised. This codon is specific to the amino acid methionine, which is nearly always the first amino acid in a polypeptide chain. At the 5’ cap of mRNA, the small 40s subunit of the ribosome binds. Subsequently, the larger 60s subunit binds to complete the initiation complex. The next step (elongation) can now commence.
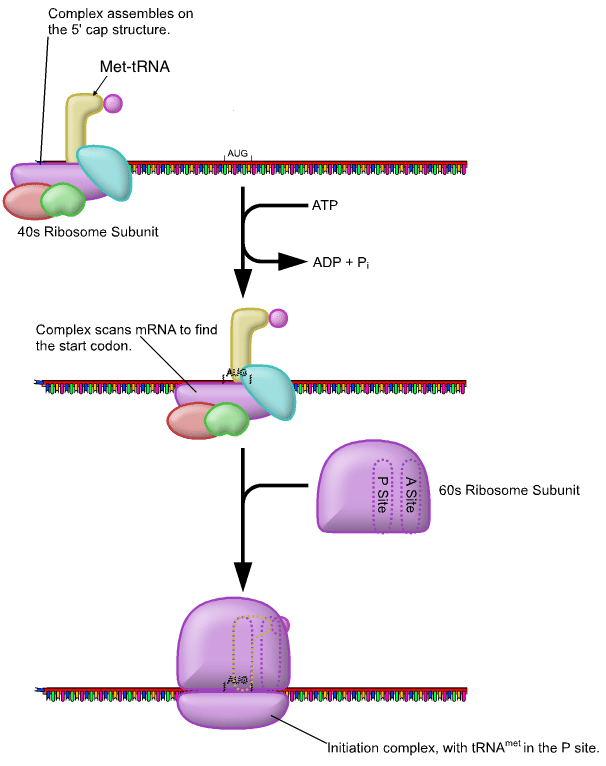
Elongation
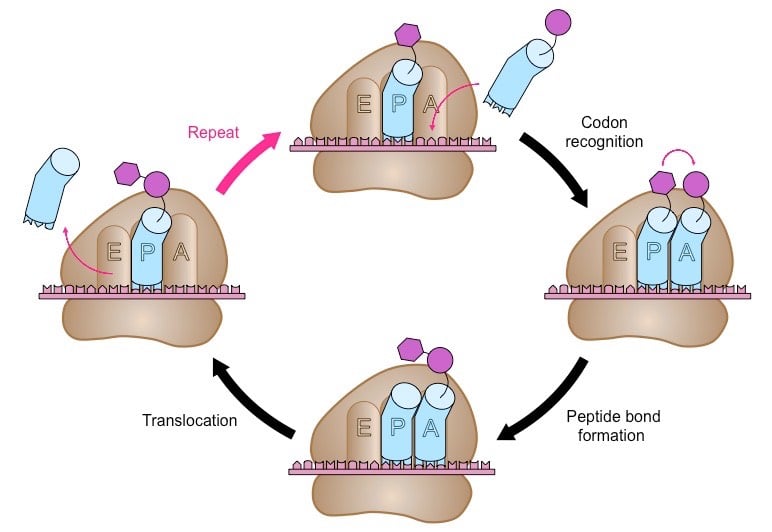
The ribosome has two tRNA binding sites; the P site which holds the peptide chain and the A site which accepts the tRNA.
While Methionine-tRNA occupies the P site, the aminoacyl-tRNA that is complementary to the next codon binds to the A site, using energy yielded from the hydrolysis of GTP.
Methionine moves from the P site to the A site to bond to a new amino acid there, starting the growth of the peptide. The tRNA molecule in the P site no longer has an attached amino acid, so leaves the ribosome.
The ribosome then translocates along the mRNA molecule to the next codon, again using energy yielded from the hydrolysis of GTP. Now, the growing peptide lies at the P site and the A site is open for the binding of the next aminoacyl-tRNA, and the cycle continues. The polypeptide chain is built up in the direction from the N terminal (methionine) to the C terminal (the final amino acid).
Termination
One of the three stop codons enters the A site. No tRNA molecules bind to these codons, so the peptide and tRNA in the P site become hydrolysed releasing the polypeptide into the cytoplasm. The small and large subunits of the ribosome dissociate, ready for the next round of translation.