PacBio Sequel IIe 16S SSU analysis - Nucleomics-VIB/16S_analysis_pipeline GitHub Wiki
(last edit: 2022-02-23)
This page explores different tools for the analysis of 16S amplicon sequencing data obtained on with conventional Illumina paired-reads or Sequel-IIE HiFi reads.
Several parameters are explored including:
- the choice of the software between several popular alternatives
- the difference in classification with the results from Illumina paired reads
This is not a complete comparison and the aim is to find a practical optimum between hands-on and result quality.
The amplicon sequencing data used in this project was published by Park et al in "Comparison of 16S rRNA Gene Based Microbial Profiling Using Five Next-Generation Sequencers and Various Primers" 1.
The paper reports results obtained from 8 different mock communities mixed from Korea registered accessions and detailed in the next tables.

Accessions
| Number | Genus | species | SILVA 138.1 with species (records) |
|---|---|---|---|
| 1 | Lactobacillus | acidophilus | (58) >Bacteria;Firmicutes;Bacilli;Lactobacillales;Lactobacillaceae;Lactobacillus;acidophilus; |
| 2* | Limosilactobacillus | fermentum | (747) >Bacteria;Firmicutes;Bacilli;Lactobacillales;Lactobacillaceae;Limosilactobacillus;NA |
| 3 | Lactobacillus | gasseri | (36) >Bacteria;Firmicutes;Bacilli;Lactobacillales;Lactobacillaceae;Lactobacillus;gasseri; |
| 4* | Lacticaseibacillus | paracasei subsp. paracasei | (587) >Bacteria;Firmicutes;Bacilli;Lactobacillales;Lactobacillaceae;Lacticaseibacillus;NA |
| 5* | Limosilactobacillus | reuteri | (747) >Bacteria;Firmicutes;Bacilli;Lactobacillales;Lactobacillaceae;Limosilactobacillus;NA |
| 6 | Bifidobacterium | animalis subsp. lactis | (96) >Bacteria;Actinobacteriota;Actinobacteria;Bifidobacteriales;Bifidobacteriaceae;Bifidobacterium;animalis; |
| 7 | Bifidobacterium | breve | (111) >Bacteria;Actinobacteriota;Actinobacteria;Bifidobacteriales;Bifidobacteriaceae;Bifidobacterium;breve; |
| 8 | Lactococcus | lactis subsp. lactis | (389) >Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Lactococcus;lactis; |
(*) records without Species level annotation
Mock communities
| Mock name | Strain | Total copy Number (copies/µL) | Ratio (%) |
|---|---|---|---|
| Mock A | Lactobacillus acidophilus | 1,40E+08 | 18,60 |
| Limosilactobacillus fermentum | 1,39E+08 | 18,45 | |
| Lactobacillus gasseri | 1,31E+08 | 17,48 | |
| Lacticaseibacillus paracasei subsp. paracasei | 1,52E+08 | 20,18 | |
| Limosilactobacillus reuteri | 1,90E+08 | 25,30 | |
| Mock B | Bifidobacterium animalis subsp. lactis | 2,58E+08 | 46,55 |
| Bifidobacterium breve | 2,96E+08 | 53,45 | |
| Mock C | Lactobacillus acidophilus | 1,39E+08 | 16,62 |
| Limosilactobacillus fermentum | 1,40E+08 | 16,79 | |
| Lactobacillus gasseri | 1,34E+08 | 15,98 | |
| Lacticaseibacillus paracasei subsp. paracasei | 1,52E+08 | 18,16 | |
| Limosilactobacillus reuteri | 1,28E+08 | 15,32 | |
| Lactococcus lactis subsp. lactis | 1,43E+08 | 17,13 | |
| Mock D | Bifidobacterium animalis subsp. lactis | 1,29E+08 | 30,73 |
| Bifidobacterium breve | 1,48E+08 | 35,14 | |
| Lactococcus lactis subsp. lactis | 1,44E+08 | 34,13 | |
| Mock E | Lactobacillus acidophilus | 6,96E+08 | 33,41 |
| Limosilactobacillus fermentum | 5,57E+08 | 26,73 | |
| Lactobacillus gasseri | 3,98E+08 | 19,12 | |
| Lacticaseibacillus paracasei subsp. paracasei | 3,03E+08 | 14,57 | |
| Limosilactobacillus reuteri | 1,28E+08 | 6,16 | |
| Mock F | Bifidobacterium animalis subsp. lactis | 3,87E+08 | 56,64 |
| Bifidobacterium breve | 2,96E+08 | 43,36 | |
| Mock G | Lactobacillus acidophilus | 3,83E+05 | 8,11 |
| Limosilactobacillus fermentum | 3,81E+05 | 8,08 | |
| Lactobacillus gasseri | 3,67E+05 | 7,78 | |
| Lacticaseibacillus paracasei subsp. paracasei. | 4,19E+05 | 8,88 | |
| Limosilactobacillus reuteri | 3,55E+05 | 7,53 | |
| Bifidobacterium animalis subsp. lactis | 8,02E+05 | 16,99 | |
| Bifidobacterium breve | 9,17E+05 | 19,43 | |
| Lactococcus lactis subsp. lactis | 1,10E+06 | 23,21 | |
| Mock H | Lactobacillus acidophilus | 1,40E+08 | 12,56 |
| Limosilactobacillus fermentum | 1,39E+08 | 12,51 | |
| Lactobacillus gasseri | 1,33E+08 | 11,96 | |
| Lacticaseibacillus paracasei subsp. paracasei | 1,52E+08 | 13,63 | |
| Limosilactobacillus reuteri | 1,29E+08 | 11,58 | |
| Bifidobacterium animalis subsp. lactis | 1,28E+08 | 11,55 | |
| Bifidobacterium breve | 1,48E+08 | 13,30 | |
| Lactococcus lactis subsp. lactis | 1,44E+08 | 12,91 |
The read data was downloaded from SRA based on the metadata table shown below and converted back to fastQ using fasterq-dump as part of NCBI SRA tools 2 .
SRA metadata
study_accession sample_accession experiment_accession run_accession tax_id scientific_name fastq_ftp submitted_ftp sra_ftp
PRJEB45207 SAMEA8898619 ERX5642659 ERR6001903 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6001903/ERR6001903.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001903/Ion_A1_1.IonCode_0170.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642660 ERR6001904 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6001904/ERR6001904.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001904/Ion_A1_2.IonCode_0171.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642661 ERR6001905 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6001905/ERR6001905.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001905/Ion_A1_3.IonCode_0172.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642662 ERR6001906 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6001906/ERR6001906.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001906/Ion_A2_1.IonCode_0201.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642663 ERR6001907 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6001907/ERR6001907.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001907/Ion_A2_2.IonCode_0202.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642664 ERR6001908 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6001908/ERR6001908.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001908/Ion_A2_3.IonCode_0203.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642665 ERR6001909 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6001909/ERR6001909.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001909/Ion_A3_1.IonCode_0233.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642666 ERR6001910 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6001910/ERR6001910.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001910/Ion_A3_2.IonCode_0234.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642667 ERR6001911 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6001911/ERR6001911.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001911/Ion_A3_3.IonCode_0235.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642668 ERR6001912 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6001912/ERR6001912.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001912/Ion_A4_1.IonCode_0265.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642669 ERR6001913 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6001913/ERR6001913.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001913/Ion_A4_2.IonCode_0266.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642670 ERR6001914 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6001914/ERR6001914.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001914/Ion_A4_3.IonCode_0267.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642671 ERR6001915 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6001915/ERR6001915.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001915/Pac_A_1.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642672 ERR6001916 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6001916/ERR6001916.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001916/Pac_A_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642673 ERR6001917 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6001917/ERR6001917.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001917/Pac_A_3.fastq.gz
PRJEB45207 SAMEA8898619 ERX5642674 ERR6001918 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6001918/ERR6001918.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6001918/MIN_A.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643144 ERR6002388 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6002388/ERR6002388.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002388/Ion_B1_2.IonCode_0174.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643145 ERR6002389 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6002389/ERR6002389.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002389/Ion_B1_3.IonCode_0175.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643146 ERR6002390 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6002390/ERR6002390.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002390/Ion_B2_1.IonCode_0204.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643147 ERR6002391 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6002391/ERR6002391.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002391/Ion_B2_2.IonCode_0205.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643148 ERR6002392 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6002392/ERR6002392.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002392/Ion_B2_3.IonCode_0206.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643149 ERR6002393 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6002393/ERR6002393.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002393/Ion_B3_1.IonCode_0236.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643150 ERR6002394 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6002394/ERR6002394.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002394/Ion_B3_2.IonCode_0237.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643151 ERR6002395 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6002395/ERR6002395.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002395/Ion_B3_3.IonCode_0238.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643152 ERR6002396 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6002396/ERR6002396.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002396/Ion_B4_1.IonCode_0268.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643153 ERR6002397 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6002397/ERR6002397.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002397/Ion_B4_2.IonCode_0269.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643154 ERR6002398 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6002398/ERR6002398.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002398/Ion_B4_3.IonCode_0270.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643155 ERR6002399 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6002399/ERR6002399.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002399/Pac_B_1.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643156 ERR6002400 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6002400/ERR6002400.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002400/Pac_B_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643157 ERR6002401 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6002401/ERR6002401.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002401/Pac_B_3.fastq.gz
PRJEB45207 SAMEA8898620 ERX5643158 ERR6002402 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6002402/ERR6002402.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002402/MIN_B.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643159 ERR6002403 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6002403/ERR6002403.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002403/Ion_C1_1.IonCode_0176.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643160 ERR6002404 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6002404/ERR6002404.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002404/Ion_C1_2.IonCode_0177.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643161 ERR6002405 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6002405/ERR6002405.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002405/Ion_C1_3.IonCode_0178.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643162 ERR6002406 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6002406/ERR6002406.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002406/Ion_C2_1.IonCode_0207.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643163 ERR6002407 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6002407/ERR6002407.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002407/Ion_C2_2.IonCode_0208.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643164 ERR6002408 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6002408/ERR6002408.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002408/Ion_C2_3.IonCode_0209.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643165 ERR6002409 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6002409/ERR6002409.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002409/Ion_C3_1.IonCode_0239.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643166 ERR6002410 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6002410/ERR6002410.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002410/Ion_C3_2.IonCode_0240.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643167 ERR6002411 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6002411/ERR6002411.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002411/Ion_C3_3.IonCode_0241.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643168 ERR6002412 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6002412/ERR6002412.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002412/Ion_C4_1.IonCode_0271.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643169 ERR6002413 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6002413/ERR6002413.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002413/Ion_C4_2.IonCode_0272.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643170 ERR6002414 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6002414/ERR6002414.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002414/Ion_C4_3.IonCode_0273.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643171 ERR6002415 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6002415/ERR6002415.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002415/Pac_C_1.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643172 ERR6002416 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6002416/ERR6002416.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002416/Pac_C_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643173 ERR6002417 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6002417/ERR6002417.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002417/Pac_C_3.fastq.gz
PRJEB45207 SAMEA8898621 ERX5643174 ERR6002418 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6002418/ERR6002418.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002418/MIN_C.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643175 ERR6002419 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6002419/ERR6002419.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002419/Ion_D1_1.IonCode_0179.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643176 ERR6002420 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6002420/ERR6002420.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002420/Ion_D1_2.IonCode_0180.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643177 ERR6002421 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6002421/ERR6002421.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002421/Ion_D1_3.IonCode_0181.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643178 ERR6002422 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6002422/ERR6002422.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002422/Ion_D2_1.IonCode_0210.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643179 ERR6002423 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6002423/ERR6002423.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002423/Ion_D2_2.IonCode_0211.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643180 ERR6002424 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6002424/ERR6002424.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002424/Ion_D2_3.IonCode_0212.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643181 ERR6002425 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6002425/ERR6002425.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002425/Ion_D3_1.IonCode_0242.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643182 ERR6002426 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6002426/ERR6002426.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002426/Ion_D3_2.IonCode_0243.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643183 ERR6002427 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6002427/ERR6002427.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002427/Ion_D3_3.IonCode_0244.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643184 ERR6002428 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6002428/ERR6002428.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002428/Ion_D4_1.IonCode_0274.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643185 ERR6002429 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6002429/ERR6002429.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002429/Ion_D4_2.IonCode_0275.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643186 ERR6002430 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6002430/ERR6002430.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002430/Ion_D4_3.IonCode_0276.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643187 ERR6002431 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6002431/ERR6002431.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002431/Pac_D_1.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643188 ERR6002432 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6002432/ERR6002432.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002432/Pac_D_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643189 ERR6002433 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6002433/ERR6002433.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002433/Pac_D_3.fastq.gz
PRJEB45207 SAMEA8898622 ERX5643190 ERR6002434 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6002434/ERR6002434.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002434/MIN_D.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643191 ERR6002435 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6002435/ERR6002435.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002435/Ion_E1_1.IonCode_0182.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643192 ERR6002436 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6002436/ERR6002436.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002436/Ion_E1_2.IonCode_0183.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643193 ERR6002437 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6002437/ERR6002437.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002437/Ion_E1_3.IonCode_0184.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643194 ERR6002438 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6002438/ERR6002438.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002438/Ion_E2_1.IonCode_0213.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643195 ERR6002439 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6002439/ERR6002439.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002439/Ion_E2_2.IonCode_0214.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643196 ERR6002440 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6002440/ERR6002440.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002440/Ion_E2_3.IonCode_0215.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643197 ERR6002441 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6002441/ERR6002441.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002441/Ion_E3_1.IonCode_0245.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643198 ERR6002442 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6002442/ERR6002442.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002442/Ion_E3_2.IonCode_0246.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643199 ERR6002443 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6002443/ERR6002443.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002443/Ion_E3_3.IonCode_0247.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643200 ERR6002444 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6002444/ERR6002444.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002444/Ion_E4_1.IonCode_0277.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643201 ERR6002445 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6002445/ERR6002445.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002445/Ion_E4_2.IonCode_0278.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643202 ERR6002446 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6002446/ERR6002446.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002446/Ion_E4_3.IonCode_0279.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643203 ERR6002447 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6002447/ERR6002447.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002447/Pac_E_1.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643204 ERR6002448 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6002448/ERR6002448.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002448/Pac_E_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643205 ERR6002449 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6002449/ERR6002449.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002449/Pac_E_3.fastq.gz
PRJEB45207 SAMEA8898623 ERX5643206 ERR6002450 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6002450/ERR6002450.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002450/MIN_E.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643207 ERR6002451 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6002451/ERR6002451.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002451/Ion_F1_1.IonCode_0185.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643208 ERR6002452 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6002452/ERR6002452.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002452/Ion_F1_2.IonCode_0186.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643209 ERR6002453 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6002453/ERR6002453.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002453/Ion_F1_3.IonCode_0187.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643210 ERR6002454 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6002454/ERR6002454.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002454/Ion_F2_1.IonCode_0216.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643211 ERR6002455 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6002455/ERR6002455.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002455/Ion_F2_2.IonCode_0217.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643212 ERR6002456 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6002456/ERR6002456.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002456/Ion_F2_3.IonCode_0218.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643213 ERR6002457 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6002457/ERR6002457.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002457/Ion_F3_1.IonCode_0248.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643214 ERR6002458 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6002458/ERR6002458.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002458/Ion_F3_2.IonCode_0249.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643215 ERR6002459 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6002459/ERR6002459.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002459/Ion_F3_3.IonCode_0250.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643216 ERR6002460 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6002460/ERR6002460.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002460/Ion_F4_1.IonCode_0280.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643217 ERR6002461 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6002461/ERR6002461.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002461/Ion_F4_2.IonCode_0281.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643218 ERR6002462 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6002462/ERR6002462.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002462/Ion_F4_3.IonCode_0282.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643219 ERR6002463 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6002463/ERR6002463.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002463/Pac_F_1.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643220 ERR6002464 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6002464/ERR6002464.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002464/Pac_F_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643221 ERR6002465 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6002465/ERR6002465.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002465/Pac_F_3.fastq.gz
PRJEB45207 SAMEA8898624 ERX5643222 ERR6002466 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6002466/ERR6002466.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002466/MIN_F.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643223 ERR6002467 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6002467/ERR6002467.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002467/Ion_G1_1.IonCode_0188.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643224 ERR6002468 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6002468/ERR6002468.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002468/Ion_G1_2.IonCode_0189.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643225 ERR6002469 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6002469/ERR6002469.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002469/Ion_G1_3.IonCode_0190.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643226 ERR6002470 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6002470/ERR6002470.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002470/Ion_G2_1.IonCode_0219.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643227 ERR6002471 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6002471/ERR6002471.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002471/Ion_G2_2.IonCode_0220.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643228 ERR6002472 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6002472/ERR6002472.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002472/Ion_G2_3.IonCode_0221.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643229 ERR6002473 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6002473/ERR6002473.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002473/Ion_G3_1.IonCode_0251.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643230 ERR6002474 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6002474/ERR6002474.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002474/Ion_G3_2.IonCode_0252.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643231 ERR6002475 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6002475/ERR6002475.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002475/Ion_G3_3.IonCode_0253.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643232 ERR6002476 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6002476/ERR6002476.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002476/Ion_G4_1.IonCode_0283.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643233 ERR6002477 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6002477/ERR6002477.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002477/Ion_G4_2.IonCode_0284.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643234 ERR6002478 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6002478/ERR6002478.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002478/Ion_G4_3.IonCode_0285.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643235 ERR6002479 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6002479/ERR6002479.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002479/Pac_G_1.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643236 ERR6002480 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6002480/ERR6002480.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002480/Pac_G_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643237 ERR6002481 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6002481/ERR6002481.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002481/Pac_G_3.fastq.gz
PRJEB45207 SAMEA8898625 ERX5643238 ERR6002482 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6002482/ERR6002482.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002482/MIN_G.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643239 ERR6002483 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6002483/ERR6002483.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002483/Ion_H1_1.IonCode_0191.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643240 ERR6002484 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6002484/ERR6002484.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002484/Ion_H1_2.IonCode_0192.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643241 ERR6002485 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6002485/ERR6002485.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002485/Ion_H1_3.IonCode_0193.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643242 ERR6002486 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6002486/ERR6002486.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002486/Ion_H2_1.IonCode_0222.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643243 ERR6002487 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6002487/ERR6002487.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002487/Ion_H2_2.IonCode_0223.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643244 ERR6002488 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6002488/ERR6002488.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002488/Ion_H2_3.IonCode_0224.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643245 ERR6002489 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6002489/ERR6002489.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002489/Ion_H3_1.IonCode_0254.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643246 ERR6002490 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6002490/ERR6002490.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002490/Ion_H3_2.IonCode_0255.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643247 ERR6002491 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6002491/ERR6002491.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002491/Ion_H3_3.IonCode_0256.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643248 ERR6002492 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6002492/ERR6002492.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002492/Ion_H4_1.IonCode_0286.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643249 ERR6002493 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6002493/ERR6002493.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002493/Ion_H4_2.IonCode_0287.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643250 ERR6002494 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6002494/ERR6002494.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002494/Ion_H4_3.IonCode_0288.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643251 ERR6002495 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6002495/ERR6002495.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002495/Pac_H_1.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643252 ERR6002496 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6002496/ERR6002496.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002496/Pac_H_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643253 ERR6002497 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6002497/ERR6002497.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002497/Pac_H_3.fastq.gz
PRJEB45207 SAMEA8898626 ERX5643254 ERR6002498 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6002498/ERR6002498.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002498/MIN_H.fastq.gz
PRJEB45207 SAMEA8898619 ERX5643255 ERR6002499 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6002499/ERR6002499.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6002499/Ion_B1_1.IonCode_0173.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644577 ERR6004017 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004017/ERR6004017_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004017/ERR6004017_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004017/MiS_A1_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004017/MiS_A1_1_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644578 ERR6004018 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004018/ERR6004018_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004018/ERR6004018_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004018/MiS_A1_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004018/MiS_A1_2_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644579 ERR6004019 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004019/ERR6004019_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004019/ERR6004019_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004019/MiS_A1_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004019/MiS_A1_3_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644580 ERR6004020 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004020/ERR6004020_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004020/ERR6004020_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004020/MiS_A2_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004020/MiS_A2_1_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644581 ERR6004021 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004021/ERR6004021_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004021/ERR6004021_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004021/MiS_A2_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004021/MiS_A2_2_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644582 ERR6004022 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004022/ERR6004022_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004022/ERR6004022_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004022/MiS_A2_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004022/MiS_A2_3_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644583 ERR6004023 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004023/ERR6004023_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004023/ERR6004023_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004023/MiS_A3_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004023/MiS_A3_1_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644584 ERR6004024 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004024/ERR6004024_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004024/ERR6004024_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004024/MiS_A3_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004024/MiS_A3_2_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644585 ERR6004025 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004025/ERR6004025_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004025/ERR6004025_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004025/MiS_A3_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004025/MiS_A3_3_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644586 ERR6004026 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004026/ERR6004026_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004026/ERR6004026_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004026/MiS_A4_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004026/MiS_A4_1_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644587 ERR6004027 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004027/ERR6004027_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004027/ERR6004027_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004027/MiS_A4_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004027/MiS_A4_2_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644588 ERR6004028 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004028/ERR6004028_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004028/ERR6004028_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004028/MiS_A4_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004028/MiS_A4_3_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644589 ERR6004029 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004029/ERR6004029_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004029/ERR6004029_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004029/MGI_A.R1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004029/MGI_A.R2_1.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644590 ERR6004030 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004030/ERR6004030_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004030/ERR6004030_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004030/MGI_A_R1_2.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004030/MGI_A_R2_2.fastq.gz
PRJEB45207 SAMEA8898619 ERX5644591 ERR6004031 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004031/ERR6004031_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004031/ERR6004031_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004031/MGI_A_R1_3.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004031/MGI_A_R2_3.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644592 ERR6004032 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004032/ERR6004032_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004032/ERR6004032_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004032/MiS_B1_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004032/MiS_B1_1_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644593 ERR6004033 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004033/ERR6004033_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004033/ERR6004033_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004033/MiS_B1_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004033/MiS_B1_2_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644594 ERR6004034 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004034/ERR6004034_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004034/ERR6004034_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004034/MiS_B1_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004034/MiS_B1_3_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644595 ERR6004035 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004035/ERR6004035_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004035/ERR6004035_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004035/MiS_B2_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004035/MiS_B2_1_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644596 ERR6004036 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004036/ERR6004036_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004036/ERR6004036_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004036/MiS_B2_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004036/MiS_B2_2_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644597 ERR6004037 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004037/ERR6004037_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004037/ERR6004037_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004037/MiS_B2_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004037/MiS_B2_3_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644598 ERR6004038 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004038/ERR6004038_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004038/ERR6004038_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004038/MiS_B3_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004038/MiS_B3_1_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644599 ERR6004039 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004039/ERR6004039_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004039/ERR6004039_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004039/MiS_B3_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004039/MiS_B3_2_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644600 ERR6004040 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004040/ERR6004040_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004040/ERR6004040_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004040/MiS_B3_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004040/MiS_B3_3_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644601 ERR6004041 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004041/ERR6004041_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004041/ERR6004041_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004041/MiS_B4_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004041/MiS_B4_1_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644602 ERR6004042 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004042/ERR6004042_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004042/ERR6004042_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004042/MIS_B4_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004042/MIS_B4_2_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644603 ERR6004043 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004043/ERR6004043_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004043/ERR6004043_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004043/MIS_B4_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004043/MIS_B4_3_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644604 ERR6004044 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004044/ERR6004044_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004044/ERR6004044_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004044/MGI_B_R1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004044/MGI_B_R2_1.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644605 ERR6004045 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004045/ERR6004045_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004045/ERR6004045_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004045/MGI_B_R1_2.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004045/MGI_B_R2_2.fastq.gz
PRJEB45207 SAMEA8898620 ERX5644606 ERR6004046 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004046/ERR6004046_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004046/ERR6004046_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004046/MGI_B_R1_3.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004046/MGI_B_R2_3.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644607 ERR6004047 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004047/ERR6004047_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004047/ERR6004047_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004047/MIS_C1_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004047/MIS_C1_1_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644608 ERR6004048 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004048/ERR6004048_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004048/ERR6004048_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004048/MIS_C1_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004048/MIS_C1_2_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644609 ERR6004049 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004049/ERR6004049_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004049/ERR6004049_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004049/MIS_C1_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004049/MIS_C1_3_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644610 ERR6004050 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004050/ERR6004050_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004050/ERR6004050_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004050/MIS_C2_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004050/MIS_C2_1_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644611 ERR6004051 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004051/ERR6004051_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004051/ERR6004051_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004051/MIS_C2_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004051/MIS_C2_2_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644612 ERR6004052 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004052/ERR6004052_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004052/ERR6004052_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004052/MIS_C2_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004052/MIS_C2_3_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644613 ERR6004053 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004053/ERR6004053_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004053/ERR6004053_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004053/MIS_C3_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004053/MIS_C3_1_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644614 ERR6004054 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004054/ERR6004054_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004054/ERR6004054_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004054/MIS_C3_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004054/MIS_C3_2_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644615 ERR6004055 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004055/ERR6004055_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004055/ERR6004055_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004055/MIS_C3_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004055/MIS_C3_3_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644616 ERR6004056 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004056/ERR6004056_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004056/ERR6004056_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004056/MIS_C4_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004056/MIS_C4_1_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644617 ERR6004057 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004057/ERR6004057_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004057/ERR6004057_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004057/MIS_C4_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004057/MIS_C4_2_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644618 ERR6004058 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004058/ERR6004058_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004058/ERR6004058_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004058/MIS_C4_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004058/MIS_C4_3_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644619 ERR6004059 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004059/ERR6004059_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004059/ERR6004059_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004059/MGI_C_R1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004059/MGI_C_R2_1.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644620 ERR6004060 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004060/ERR6004060_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004060/ERR6004060_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004060/MGI_C_R1_2.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004060/MGI_C_R2_2.fastq.gz
PRJEB45207 SAMEA8898621 ERX5644621 ERR6004061 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004061/ERR6004061_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004061/ERR6004061_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004061/MGI_C_R1_3.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004061/MGI_C_R2_3.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644622 ERR6004062 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004062/ERR6004062_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004062/ERR6004062_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004062/MIS_D1_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004062/MIS_D1_1_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644623 ERR6004063 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004063/ERR6004063_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004063/ERR6004063_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004063/MIS_D1_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004063/MIS_D1_2_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644624 ERR6004064 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004064/ERR6004064_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004064/ERR6004064_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004064/MIS_D1_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004064/MIS_D1_3_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644625 ERR6004065 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004065/ERR6004065_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004065/ERR6004065_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004065/MIS_D2_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004065/MIS_D2_1_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644626 ERR6004066 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004066/ERR6004066_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004066/ERR6004066_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004066/MIS_D2_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004066/MIS_D2_2_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644627 ERR6004067 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004067/ERR6004067_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004067/ERR6004067_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004067/MIS_D2_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004067/MIS_D2_3_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644628 ERR6004068 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004068/ERR6004068_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004068/ERR6004068_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004068/MIS_D3_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004068/MIS_D3_1_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644629 ERR6004069 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004069/ERR6004069_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004069/ERR6004069_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004069/MIS_D3_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004069/MIS_D3_2_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644630 ERR6004070 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004070/ERR6004070_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004070/ERR6004070_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004070/MIS_D3_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004070/MIS_D3_3_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644631 ERR6004071 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004071/ERR6004071_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004071/ERR6004071_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004071/MIS_D4_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004071/MIS_D4_1_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644632 ERR6004072 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004072/ERR6004072_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004072/ERR6004072_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004072/MIS_D4_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004072/MIS_D4_2_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644633 ERR6004073 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004073/ERR6004073_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004073/ERR6004073_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004073/MIS_D4_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004073/MIS_D4_3_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644634 ERR6004074 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004074/ERR6004074_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004074/ERR6004074_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004074/MGI_D_R1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004074/MGI_D_R2_1.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644635 ERR6004075 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004075/ERR6004075_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004075/ERR6004075_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004075/MGI_D_R1_2.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004075/MGI_D_R2_2.fastq.gz
PRJEB45207 SAMEA8898622 ERX5644636 ERR6004076 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004076/ERR6004076_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004076/ERR6004076_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004076/MGI_D_R1_3.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004076/MGI_D_R2_3.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644637 ERR6004077 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004077/ERR6004077_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004077/ERR6004077_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004077/MIS_E1_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004077/MIS_E1_1_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644638 ERR6004078 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004078/ERR6004078_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004078/ERR6004078_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004078/MIS_E1_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004078/MIS_E1_2_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644639 ERR6004079 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004079/ERR6004079_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004079/ERR6004079_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004079/MIS_E1_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004079/MIS_E1_3_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644640 ERR6004080 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004080/ERR6004080_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004080/ERR6004080_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004080/MIS_E2_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004080/MIS_E2_1_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644641 ERR6004081 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004081/ERR6004081_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004081/ERR6004081_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004081/MIS_E2_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004081/MIS_E2_2_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644642 ERR6004082 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004082/ERR6004082_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004082/ERR6004082_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004082/MIS_E2_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004082/MIS_E2_3_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644643 ERR6004083 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004083/ERR6004083_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004083/ERR6004083_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004083/MIS_E3_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004083/MIS_E3_1_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644644 ERR6004084 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004084/ERR6004084_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004084/ERR6004084_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004084/MIS_E3_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004084/MIS_E3_2_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644645 ERR6004085 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004085/ERR6004085_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004085/ERR6004085_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004085/MIS_E3_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004085/MIS_E3_3_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644646 ERR6004086 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004086/ERR6004086_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004086/ERR6004086_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004086/MIS_E4_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004086/MIS_E4_1_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644647 ERR6004087 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004087/ERR6004087_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004087/ERR6004087_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004087/MIS_E4_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004087/MIS_E4_2_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644648 ERR6004088 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004088/ERR6004088_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004088/ERR6004088_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004088/MIS_E4_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004088/MIS_E4_3_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644649 ERR6004089 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004089/ERR6004089_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004089/ERR6004089_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004089/MGI_E_R1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004089/MGI_E_R2_1.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644650 ERR6004090 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004090/ERR6004090_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004090/ERR6004090_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004090/MGI_E_R1_2.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004090/MGI_E_R2_2.fastq.gz
PRJEB45207 SAMEA8898623 ERX5644651 ERR6004091 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004091/ERR6004091_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004091/ERR6004091_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004091/MGI_E_R1_3.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004091/MGI_E_R2_3.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644652 ERR6004092 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004092/ERR6004092_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004092/ERR6004092_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004092/MIS_F1_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004092/MIS_F1_1_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644653 ERR6004093 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004093/ERR6004093_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004093/ERR6004093_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004093/MIS_F1_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004093/MIS_F1_2_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644654 ERR6004094 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004094/ERR6004094_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004094/ERR6004094_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004094/MIS_F1_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004094/MIS_F1_3_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644655 ERR6004095 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004095/ERR6004095_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004095/ERR6004095_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004095/MIS_F2_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004095/MIS_F2_1_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644656 ERR6004096 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004096/ERR6004096_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004096/ERR6004096_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004096/MIS_F2_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004096/MIS_F2_2_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644657 ERR6004097 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004097/ERR6004097_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004097/ERR6004097_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004097/MIS_F2_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004097/MIS_F2_3_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644658 ERR6004098 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004098/ERR6004098_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004098/ERR6004098_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004098/MIS_F3_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004098/MIS_F3_1_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644659 ERR6004099 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004099/ERR6004099_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004099/ERR6004099_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004099/MIS_F3_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004099/MIS_F3_2_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644660 ERR6004100 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004100/ERR6004100_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004100/ERR6004100_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004100/MIS_F3_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004100/MIS_F3_3_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644661 ERR6004101 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004101/ERR6004101_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004101/ERR6004101_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004101/MIS_F4_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004101/MIS_F4_1_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644662 ERR6004102 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004102/ERR6004102_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004102/ERR6004102_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004102/MIS_F4_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004102/MIS_F4_2_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644663 ERR6004103 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004103/ERR6004103_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004103/ERR6004103_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004103/MIS_F4_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004103/MIS_F4_3_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644664 ERR6004104 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004104/ERR6004104_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004104/ERR6004104_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004104/MGI_F_R1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004104/MGI_F_R2_1.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644665 ERR6004105 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004105/ERR6004105_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004105/ERR6004105_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004105/MGI_F_R1_2.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004105/MGI_F_R2_2.fastq.gz
PRJEB45207 SAMEA8898624 ERX5644666 ERR6004106 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004106/ERR6004106_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004106/ERR6004106_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004106/MGI_F_R1_3.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004106/MGI_F_R2_3.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644667 ERR6004107 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004107/ERR6004107_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004107/ERR6004107_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004107/MIS_G1_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004107/MIS_G1_1_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644668 ERR6004108 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004108/ERR6004108_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004108/ERR6004108_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004108/MIS_G1_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004108/MIS_G1_2_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644669 ERR6004109 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004109/ERR6004109_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004109/ERR6004109_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004109/MIS_G1_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004109/MIS_G1_3_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644670 ERR6004110 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004110/ERR6004110_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004110/ERR6004110_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004110/MIS_G2_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004110/MIS_G2_1_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644671 ERR6004111 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004111/ERR6004111_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004111/ERR6004111_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004111/MIS_G2_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004111/MIS_G2_2_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644672 ERR6004112 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004112/ERR6004112_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004112/ERR6004112_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004112/MIS_G2_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004112/MIS_G2_3_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644673 ERR6004113 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004113/ERR6004113_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004113/ERR6004113_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004113/MIS_G3_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004113/MIS_G3_1_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644674 ERR6004114 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004114/ERR6004114_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004114/ERR6004114_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004114/MIS_G3_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004114/MIS_G3_2_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644675 ERR6004115 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004115/ERR6004115_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004115/ERR6004115_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004115/MIS_G3_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004115/MIS_G3_3_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644676 ERR6004116 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004116/ERR6004116_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004116/ERR6004116_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004116/MIS_G4_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004116/MIS_G4_1_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644677 ERR6004117 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004117/ERR6004117_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004117/ERR6004117_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004117/MIS_G4_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004117/MIS_G4_2_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644678 ERR6004118 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004118/ERR6004118_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004118/ERR6004118_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004118/MIS_G4_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004118/MIS_G4_3_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644679 ERR6004119 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004119/ERR6004119_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004119/ERR6004119_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004119/MGI_G_R1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004119/MGI_G_R2_1.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644680 ERR6004120 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004120/ERR6004120_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004120/ERR6004120_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004120/MGI_G_R1_2.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004120/MGI_G_R2_2.fastq.gz
PRJEB45207 SAMEA8898625 ERX5644681 ERR6004121 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004121/ERR6004121_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004121/ERR6004121_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004121/MGI_G_R1_3.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004121/MGI_G_R2_3.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644682 ERR6004122 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004122/ERR6004122_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004122/ERR6004122_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004122/MIS_H1_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004122/MIS_H1_1_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644683 ERR6004123 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004123/ERR6004123_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004123/ERR6004123_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004123/MIS_H1_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004123/MIS_H1_2_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644684 ERR6004124 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004124/ERR6004124_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004124/ERR6004124_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004124/MIS_H1_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004124/MIS_H1_3_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644685 ERR6004125 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004125/ERR6004125_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004125/ERR6004125_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004125/MIS_H2_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004125/MIS_H2_1_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644686 ERR6004126 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004126/ERR6004126_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004126/ERR6004126_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004126/MIS_H2_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004126/MIS_H2_2_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644687 ERR6004127 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004127/ERR6004127_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/007/ERR6004127/ERR6004127_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004127/MIS_H2_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004127/MIS_H2_3_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644688 ERR6004128 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004128/ERR6004128_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/008/ERR6004128/ERR6004128_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004128/MIS_H3_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004128/MIS_H3_1_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644689 ERR6004129 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004129/ERR6004129_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/009/ERR6004129/ERR6004129_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004129/MIS_H3_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004129/MIS_H3_2_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644690 ERR6004130 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004130/ERR6004130_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/000/ERR6004130/ERR6004130_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004130/MIS_H3_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004130/MIS_H3_3_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644691 ERR6004131 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004131/ERR6004131_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/001/ERR6004131/ERR6004131_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004131/MIS_H4_1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004131/MIS_H4_1_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644692 ERR6004132 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004132/ERR6004132_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/002/ERR6004132/ERR6004132_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004132/MIS_H4_2_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004132/MIS_H4_2_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644693 ERR6004133 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004133/ERR6004133_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/003/ERR6004133/ERR6004133_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004133/MiS_H4_3_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004133/MiS_H4_3_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644694 ERR6004134 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004134/ERR6004134_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/004/ERR6004134/ERR6004134_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004134/MGI_H_R1_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004134/MGI_H_R2_1.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644695 ERR6004135 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004135/ERR6004135_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/005/ERR6004135/ERR6004135_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004135/MGI_H_R1_2.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004135/MGI_H_R2_2.fastq.gz
PRJEB45207 SAMEA8898626 ERX5644696 ERR6004136 1869227 bacterium ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004136/ERR6004136_1.fastq.gz;ftp.sra.ebi.ac.uk/vol1/fastq/ERR600/006/ERR6004136/ERR6004136_2.fastq.gz ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004136/MGI_H_R1_3.fastq.gz;ftp.sra.ebi.ac.uk/vol1/run/ERR600/ERR6004136/MGI_H_R2_3.fastq.gz
Several published tools were selected based on their use with PacBio long read amplicons.
The tools selected here all use DADA2 for the processing of the amplicon data and SILVA as database for the classification of the obtained OTU (Operational taxonomic unit) or ASV (amplicon sequence variant) entities. The latest version of SILVA (v138.1) 3 was chosen for classification as it represents the mosty up-to-date normalized set of bacterial references.
Note: A OTU is often associated with s single organism, this is not necessarily true for an ASV which could correspond to a specific 16S copy of a given organism, provided most bacteria (and likely other organisms too) have multiple distinct copies of the rRNA genes, each harboring sequence variations (hence the name ASV). For this reason, summing up the ASV copies will not give the count of individual organism but instead create counts larger than the truth.
The DADA2 Bioconductor package (v1.22) is an improved version of the original DADA package and was published by Callahan et al in 2016 4.
We ran a selection of commands taken from the online DADA2 tutorial (v1.16) posted on github.io 5
The DADA2 analysis of Pacbio V1V9 reads was published by Callaghan et al 4b and a tutorial is available online
(snakemake workflow)
The dadasnake snakemake workflow 6 is based on DADA2 with addition of QC and annotation steps and performs additional tasks like computing of a tree and classification using several optional tools (dada2, mothur, decipher, blast). We did not perform all possible analyses with the pipeline in order to obtain data comparable with the other tools.
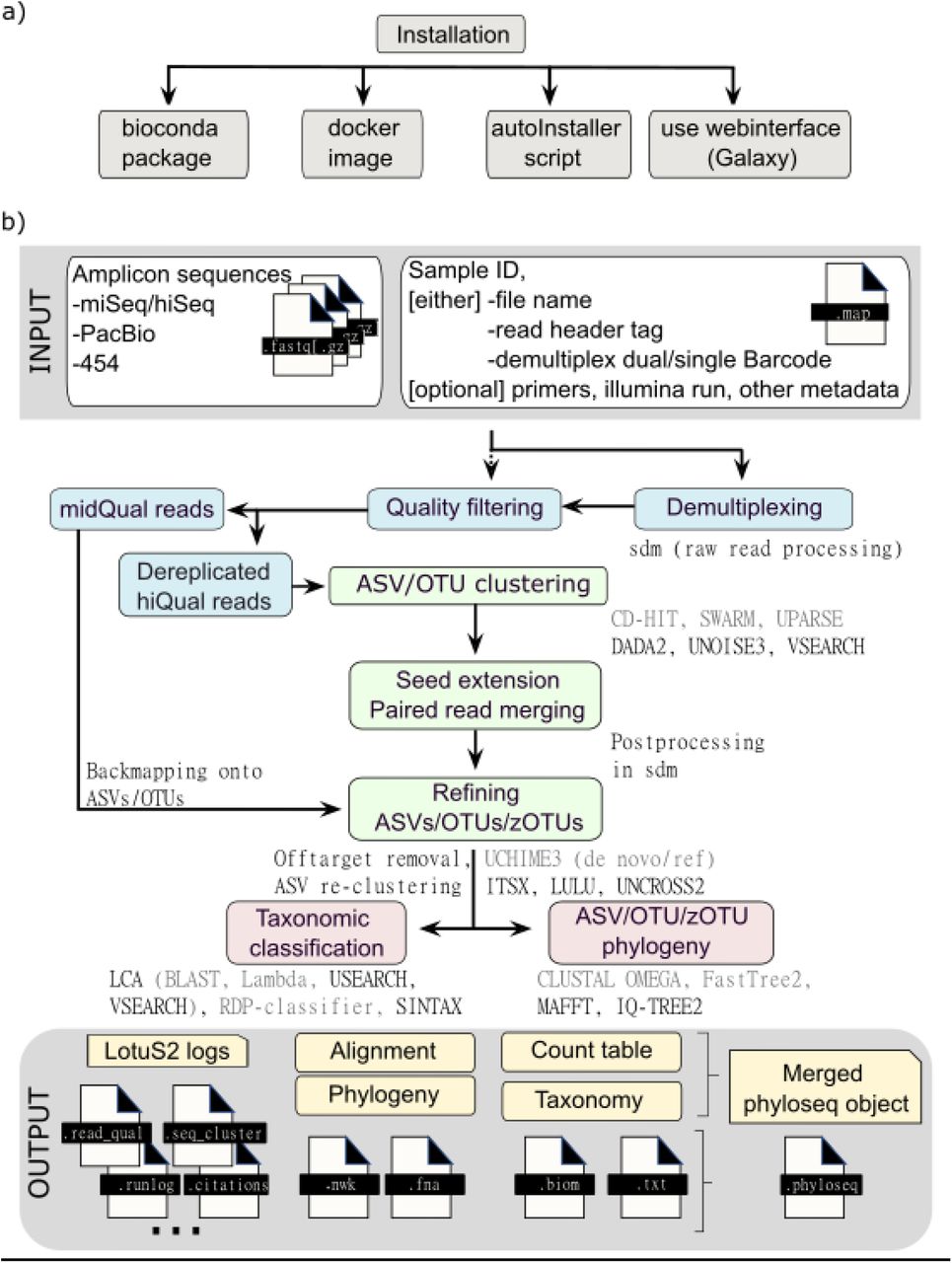
Similarly to dadasnake, the Lotus2 tool 7 is based on DADA2 for the core analysis but adds extra steps to better classify the data. It can be run on a cluster or on a server (done here) and can scale to a large number of sample (>10'000 reported by the authors).
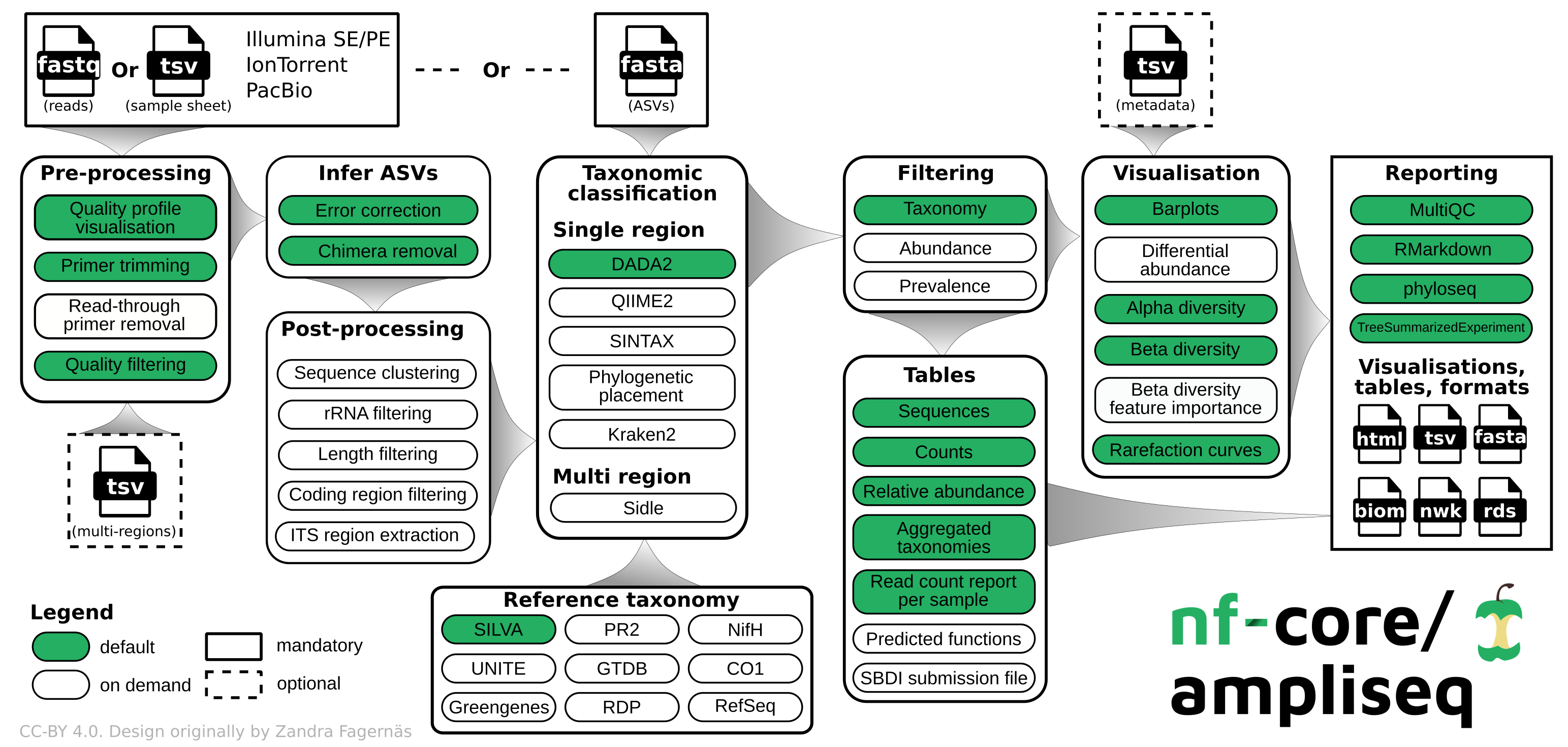
The Nextflow workflow Ampliseq 8 is only cited here for completeness and will be evaluated at a later point as it uses QIIME2 which is broadly used for Illumina data and not yet validated for long-read analysis (although it should be applicable too).
The FASAS workflow 8 was recently published for the analysis of de-novo assembled full length 16S from short reads to complete 16S loci and binning the resulting sequences. Although not specifically applied to long reads, this method could probably be inspired from and adapted to SequelIIe HiFi reads.

1: Park et al, 2021 (doi) ↩
2: NCBI SRA tools on github (link) ↩
3: SILVA database v138.1 (link) ↩
4: Callahan et al, 2016 (doi) ↩
5: DADA2 v1.16 github.io tutorial (link) ↩
4b: Callahan et al, 2019 (doi) ↩
6: Weißbecker et al, 2020 (doi) ↩
7: Özkurt et al, 2021 (doi, link) ↩
8: Straub et al, 2020 (doi, link) ↩