MRI_Spot_Coloc_Tool - MontpellierRessourcesImagerie/imagej_macros_and_scripts GitHub Wiki
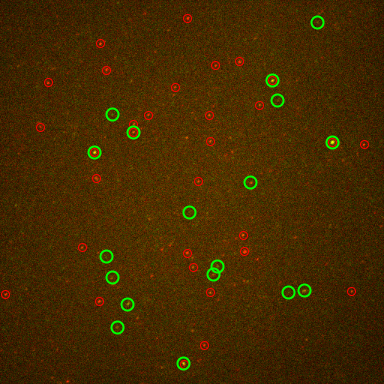
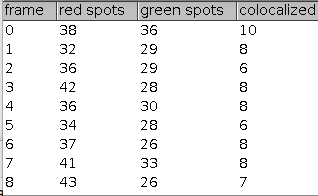
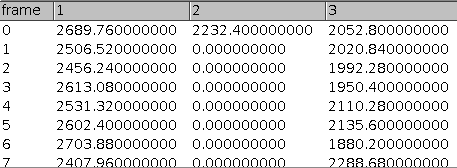 The tool counts the number of co-localized spots in two channels over time. It also exports the intensity of each spot over time.
The tool counts the number of co-localized spots in two channels over time. It also exports the intensity of each spot over time.
You can download an example image here. You will need to change the prominence values for this example.
The source code in git-hub can be found here.
Getting started
You must have FeatureJ installed.
To install the tool save the file mri_spot_coloc_tool.ijm into the folder macros/toolsets of your FIJI installation.
Select the "mri_spot_coloc_tool" toolset from the >> button of the ImageJ launcher.

Open a two channel time series image and press the d-button.
Options
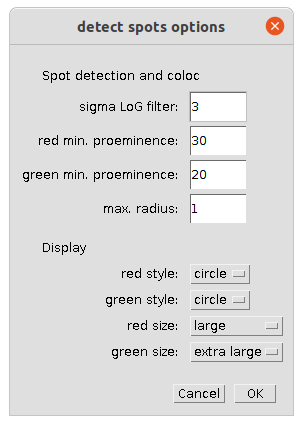
- sigma LoG filter: The sigma of the laplacian of gaussian filter (depends on the size of the spots)
- red min. prominence: The minimum prominence of the minima for the spots in the red channel (channel 1)
- green min. prominence: The minimum prominence of the minima for the spots in the green channel (channel 2)
- max. radius: spots closer to each other than 2*(radius + 1) pixel are considered co-localized.
- red style: the style in which the detected red spots are displayed.
- green style: the style in which the detected green spots are displayed.
- red size: the size of the roi marking a red spot.
- green size: the size of the roi marking a green spot.
Method
FeatureJ is used to apply a LoG filter on each channel. The spots are detected as minima with the given prominence in each channel and on each frame. Spots overlapping with the given max. radius are connected across neighbouring frames. Red and green spots overlapping with respect to the given max. radius are counted as co-localized for each frame.