Intensity Ratio Nuclei Cytoplasm Tool - MontpellierRessourcesImagerie/imagej_macros_and_scripts GitHub Wiki
The tool calculates the ratio of the intensity in the nuclei and the cytoplasm. It needs two images as input: the cytoplasm channel and the nuclei channel. The nuclei channel is used to segment the nuclei. The measurements are made in the cytoplasm channel after the background intensity has been corrected.
Getting started
To install the tools, save the file Intensity_Ratio_Nuclei_Cytoplasm.ijm inti the macros/toolsets folder of your ImageJ installation.
Select the "Intensity_Ratio_Nuclei_Cytoplasm" toolset from the >> button of the ImageJ launcher.

- the first button (the one with the image) opens this help page
- the c-button runs the correct background method on the current image
- the s-button measures the the intensity ratio for a single image (the second channel is loaded automatically)
- the b-button runs the tool in batch mode on a given folder
Background correction
Open an image (cytoplasm channel) and press the c-button. The image will become black and after a moment the correction is done. Zero values will be displayed in blue. Everything displayed in grey will be counted as cytoplasm intensity later.
The operation will search the minimum value in the image and than take into account all pixels with intensities smaller than the minimum value in the image plus the offset. For all these pixels it will search the biggest value in the neighbourhood defined by the radius option. The biggest value found in one of those neighbourhoods will than be subtracted from the image.
Right click on the c-button to modify the options of the operation:
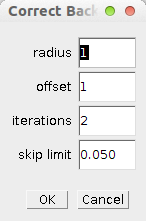
- radius - the radius of the neighbourhood in which the max. value near a minimum is searched.
- offset - the maximum is searches around pixels with an intensity smaller than the global minimum plus the offset
- iterations - the number of times the procedure is repeated
- skip limit - the ratio of pixels with intensity zero in the image above which the operation is skipped to avoid subtracting from images for which the background is already zero
Intensity ratio for single image
Open an image and press the s-button. The corresponding second channel will be loaded and the intensity ratio between the intensity within the nuclei and the cytoplasm will be measured.
A right click on the s-button (or the b-button) opens the options-dialog:
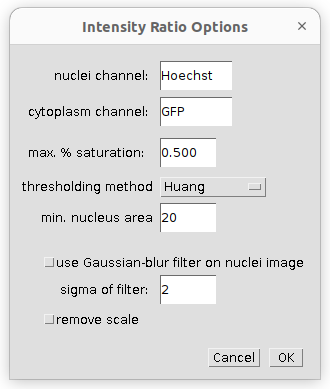
- nuclei channel - name of the staining for the nuclei channel (must be part of the filename)
- cytoplasm channel - name of the staining of the cytoplasm channel (must be part of the filename and the rest of the filename must be the same as for the nuclei image).
- max. % saturation - maximal percentage of saturated pixels allowed for the image to be taken into account
- thresholding method - the thresholding method used to segment the nuclei
- min. nucleus area - when selecting the nuclei, objects smaller than the min. area will not be taken into account
- use Gaussian-blur filter on nuclei image - if selected a Gaussian-blur filter is applied to the nuclei image, before the nuclei are segmented
- sigma of filter - the sigma of the Gaussian-blur filter
- remove scale - if selected the spatial calibration of the image (if any) is removed, so that the area measurements are in pixel.
Intensity ratio batch
Click on the b-button. A file-dialog will be opened. Browse to the folder containing the input images and select it. The progress will be displayed in a log window. When the batch processing finished you can find a file "INTENSITY-RATIO.xls" containing the result measurements in the input folder. A subfolder containing the control images has been created in the input folder as well.
Results

The measured result values are:
- image - the name of the image
- icn factor - the factor between the cytoplasm and the nuclei intensity (%nuclei = %cytoplasm * icn)
- % nuclei - percentage of the total intensity in the nuclei area
- % cytoplasm - percentage of the total intensity in the cytoplasm area
- av. nuclei intensity - the average intensity in the nuclei area
- av. cytoplasm intensity - the average intensity in the cytoplasm area
- nuclei area - the surface of the nuclei area (in pixel or in the scale set in the image)
- cytoplasm area - the surface of the cytoplasm area (in pixel or in the scale set in the image)
- t. nuclei intensity - the total intensity in the nuclei area (sum of all the intensity values)
- t. cytoplasm intensity - the total intensity in the cytoplasm area (sum of all the intensity values)
- folder - the folder from which the image was loaded
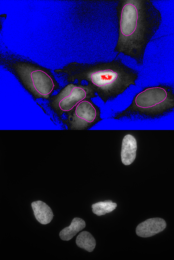
The control image shows the segmented nuclei and the cytoplasm area taken into account:
How to cite the tool
You can use the tool's resource id (see scicrunch.org):
(Intensity Ratio Nuclei Cytoplasm Tool, RRID:SCR_018573)
Publications using the tool
-
Tran, S.K., Lichtenberg, J.Y., Leonard, C.E., Williamson, J.R., Sterling, H.R., Panek, G.K., Pearson, A.H., Lopez, S., Lemmon, C.A., Conway, D.E., Hwang, P.Y., 2025. P-cadherin-dependent adhesions are required for single lumen formation and HGF-mediated cell protrusions during epithelial morphogenesis. iScience 28, 111844.
-
Schuhknecht, L., Ortmayr, K., Jänes, J., Bläsi, M., Panoussis, E., Bors, S., Dorčáková, T., Fuhrer, T., Beltrao, P., Zampieri, M., 2025. A human metabolic map of pharmacological perturbations reveals drug modes of action. Nat Biotechnol.
-
Friedman, R.M., Truong, H.D., Aronson, M.R., Brown, E.A., Angelozzi, M., Chen, J.F., Zur, K.B., Lefebvre, V., Gottardi, R., 2025. Inhibition of the MRTF-A/SRF signaling axis alleviates vocal fold scarring. Matrix Biol 137, 1–11.
-
Gerardo, H., Lourenço, T., Torres, J., Ferreira, M., Aveleira, C., Simões, S., Ferreira, L., Cavadas, C., Oliveira, P.J., Teixeira, J., Grãos, M., 2025. Extracellular matrix mechanical cues (dys)regulate metabolic redox homeostasis due to impaired autophagic flux. Eur J Clin Invest 55, e70051.
-
Sztachera, M., Wendlandt-Stanek, W., Serwa, R.A., Stanaszek, L., Smuszkiewicz, M., Wronka, D., Piwecka, M., 2025. Interrogation of RNA-bound proteome with XRNAX illuminates molecular alterations in the mouse brain affected with dysmyelination. Cell Reports 44, 115095. https://doi.org/10.1016/j.celrep.2024.115095
-
Passi, M., Stöckl, J.B., Fröhlich, T., Moser, S., Vollmar, A.M., Zahler, S., 2024. CDK5 interacts with MST2 and modulates the Hippo signalling pathway. FEBS Open Bio 2211-5463.13962.
-
Iwanski, J.B., Pappas, C.T., Mayfield, R.M., Farman, G.P., Ahrens-Nicklas, R., Churko, J.M., Gregorio, C.C., 2024. Leiomodin 2 neonatal dilated cardiomyopathy mutation results in altered actin gene signatures and cardiomyocyte dysfunction. npj Regen Med 9, 21. 10.1038/s41536-024-00366-y
-
Marugán, C., Sanz‐Gómez, N., Ortigosa, B., Monfort‐Vengut, A., Bertinetti, C., Teijo, A., González, M., Alonso De La Vega, A., Lallena, M.J., Moreno‐Bueno, G., De Cárcer, G., 2024. TPX2 overexpression promotes sensitivity to dasatinib in breast cancer by activating YAP transcriptional signaling. Molecular Oncology 18, 1531–1551. 10.1002/1878-0261.13602
-
Bougon, J., Kadijk, E., Gallot-Lavallee, L., Curtis, B.A., Landers, M., Archibald, J.M., Khaperskyy, D.A., 2024. Influenza A virus NS1 effector domain is required for PA-X-mediated host shutoff in infected cells. J Virol 98, e01901-23. 10.1128/jvi.01901-23
-
Papadimitriou, L., Karagiannaki, A., Stratakis, E., and Ranella, A. (2024). Substrate topography affects PC12 cell differentiation through mechanotransduction mechanisms. Mechanobiology in Medicine 2, 100039. 10.1016/j.mbm.2024.100039.
-
Prekovic, S., Chalkiadakis, T., Roest, M., Roden, D., Lutz, C., Schuurman, K., Opdam, M., Hoekman, L., Abbott, N., Tesselaar, T., et al. (2023). Luminal breast cancer identity is determined by loss of glucocorticoid receptor activity. EMBO Mol Med 15, e17737. 10.15252/emmm.202317737.
-
Li, Y., Shah, R.B., Sarti, S., Belcher, A.L., Lee, B.J., Gorbatenko, A., Nemati, F., Yu, H., Stanley, Z., Rahman, M., et al. (2023). A noncanonical IRAK4-IRAK1 pathway counters DNA damage–induced apoptosis independently of TLR/IL-1R signaling. Sci. Signal. 16, eadh3449. 10.1126/scisignal.adh3449.
-
Claude-Taupin, A., Isnard, P., Bagattin, A., Kuperwasser, N., Roccio, F., Ruscica, B., Goudin, N., Garfa-Traoré, M., Regnier, A., Turinsky, L., et al. (2023). The AMPK-Sirtuin 1-YAP axis is regulated by fluid flow intensity and controls autophagy flux in kidney epithelial cells. Nat Commun 14, 8056. 10.1038/s41467-023-43775-1.
-
Mathai, C., Jourd’heuil, F., Pham, L.G.C., Gilliard, K., Howard, D., Balnis, J., Jaitovich, A., Chittur, S.V., Rilley, M., Peredo-Wende, R., et al. (2023). Regulation of DNA damage and transcriptional output in the vasculature through a cytoglobin-HMGB2 axis. Redox Biology 65, 102838. 10.1016/j.redox.2023.102838.
-
Vélez, E.J., Schnebert, S., Goguet, M., Balbuena-Pecino, S., Dias, K., Beauclair, L., Fontagné-Dicharry, S., Véron, V., Depincé, A., Beaumatin, F., et al. (2023). Chaperone-mediated autophagy protects against hyperglycemic stress. Autophagy, 1–17. 10.1080/15548627.2023.2267415.
-
Messelodi, D., Strocchi, S., Bertuccio, S.N., Baden, P., Indio, V., Giorgi, F.M., Taddia, A., Serravalle, S., Valente, S., Di Fonzo, A., et al. (2023). Neuronopathic Gaucher disease models reveal defects in cell growth promoted by Hippo pathway activation. Commun Biol 6, 431. 10.1038/s42003-023-04813-2.
-
Mehak Passi (2023). CDK5 interacts with STK3- A consequence to the Hippo Signaling.
-
Richard Kondwani Maganga, An investigation of the mechanisms underlying the developmental origins of health and disease and their impact on telomeres, University of Western Australia, Perth, 2023.
-
Kretschmer, M., Mamistvalov, R., Sprinzak, D., Vollmar, A.M., and Zahler, S. (2023). Matrix stiffness regulates Notch signaling activity in endothelial cells. Journal of Cell Science 136, jcs260442. 10.1242/jcs.260442.
-
Mehak Passi, CDK5 interacts with STK3- A consequence to the Hippo Signaling, Ludwig-Maximilians-Universität, München, 2023.
-
Dincã, D. M. et al. Myotonic dystrophy RNA toxicity alters morphology, adhesion and migration of mouse and human astrocytes. Nat Commun 13, 3841 (2022).
-
Li, H., Duann, P., Li, Z., Zhou, X., Ma, J., Rovin, B.H., Lin, P.-H., 2022. The cell membrane repair protein MG53 modulates transcription factor NF-κB signaling to control kidney fibrosis. Kidney Int 101, 119–130.
-
Tak, Y.J., and Kang, S. (2022). The E2 ubiquitin-conjugating enzyme HIP2 is a crucial regulator of quality control against mutant SOD1 proteotoxicity. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1868, 166316. 10.1016/j.bbadis.2021.166316.
-
Liu, Q. et al. Hypoxia Triggers TAZ Phosphorylation in Basal A Triple Negative Breast Cancer Cells. IJMS 23, 10119 (2022).
-
Guillon, A. et al. Host succinate inhibits influenza virus infection through succinylation and nuclear retention of the viral nucleoprotein. The EMBO Journal 41, (2022).
-
Ghosh, S., Leng, W., Wilsch-Bräuninger, M., Barrera-Velázquez, M., Léopold, P., and Eaton, S. (2022). A local insulin reservoir in Drosophila alpha cell homologs ensures developmental progression under nutrient shortage. Current Biology 32, 1788-1797.e5. 10.1016/j.cub.2022.02.068.
-
Hurth, Z., Faber, M.-L., Gendrisch, F., Holzer, M., Haarhaus, B., Cawelius, A., Schwabe, K., Schempp, C.M., Wölfle, U., 2022. The Anti-Inflammatory Effect of Humulus lupulus Extract In Vivo Depends on the Galenic System of the Topical Formulation. Pharmaceuticals 15, 350. https://doi.org/10.3390/ph15030350
-
Daghero, H. et al. Jejunum-derived NF-κB reporter organoids as 3D models for the study of TNF-alpha-induced inflammation. Sci Rep 12, 14425 (2022).
-
Matthew T. Sacco, Cytoplasmic N6-Methyladenosine Deposition on Hepatitis C Viral RNA, Duke University, Durham, 2022.
-
Cai, W., Srivastava, P., Feng, D., Lin, Y., Vanderburg, C.R., Xu, Y., Mclean, P., Frosch, M.P., Fisher, D.E., Schwarzschild, M.A., et al. (2022). Melanocortin 1 receptor activation protects against alpha-synuclein pathologies in models of Parkinson’s disease. Mol Neurodegeneration 17, 16. 10.1186/s13024-022-00520-4.
-
Han P, Guo T, Jayasree A, et al. Tunable Nano-engineered Anisotropic Surface for Enhanced Mechanotransduction and Soft-Tissue Integration. Nano Research, 2022, https://doi.org/10.1007/s12274-023-5379-y
-
Sacco, M.T., Bland, K.M., and Horner, S.M. (2022). WTAP Targets the METTL3 m 6 A-Methyltransferase Complex to Cytoplasmic Hepatitis C Virus RNA to Regulate Infection. J Virol 96, e00997-22. 10.1128/jvi.00997-22.
-
Fort, L., Gama, V., and Macara, I.G. (2022). Stem cell conversion to the cardiac lineage requires nucleotide signalling from apoptosing cells. Nat Cell Biol 24, 434–447. 10.1038/s41556-022-00888-x.
-
Kaur, N., Ruiz-Velasco, A., Raja, R., Howell, G., Miller, J.M., Abouleisa, R.R.E., Ou, Q., Mace, K., Hille, S.S., Frey, N., Binder, P., Smith, C.P., Fachim, H., Soran, H., Swanton, E., Mohamed, T.M.A., Müller, O.J., Wang, X., Chernoff, J., Cartwright, E.J., Liu, W., 2022. Paracrine signal emanating from stressed cardiomyocytes aggravates inflammatory microenvironment in diabetic cardiomyopathy. iScience 25, 103973. https://doi.org/10.1016/j.isci.2022.103973
-
Kadiri, M., Charbonneau, M., Lalanne, C., Harper, K., Balg, F., Marotta, A., Dubois, C.M., 2021. 14-3-3η Promotes Invadosome Formation via the FOXO3–Snail Axis in Rheumatoid Arthritis Fibroblast-like Synoviocytes. IJMS 23, 123. https://doi.org/10.3390/ijms23010123
-
Zhu, C., Kim, S.-J., Mooradian, A., Wang, F., Li, Z., Holohan, S., Collins, P.L., Wang, K., Guo, Z., Hoog, J., Ma, C.X., Oltz, E.M., Held, J.M., Shao, J., 2021. Cancer-associated exportin-6 upregulation inhibits the transcriptionally repressive and anticancer effects of nuclear profilin-1. Cell Reports 34, 108749. 10.1016/j.celrep.2021.108749
-
Hsu, C. G., Fazal, F., Rahman, A., Berk, B. C. & Yan, C. Phosphodiesterase 10A Is a Key Mediator of Lung Inflammation. J.I. 206, 3010–3020 (2021).
-
Payapilly, A. et al. TIAM1-RAC1 promote small-cell lung cancer cell survival through antagonizing Nur77-induced BCL2 conformational change. Cell Reports 37, 109979 (2021).
-
Ramic, M. et al. Epigenetic Small Molecules Rescue Nucleocytoplasmic Transport and DNA Damage Phenotypes in C9ORF72 ALS/FTD. Brain Sciences 11, 1543 (2021).
-
Pablos, I. et al. Mechanistic insights into COVID-19 by global analysis of the SARS-CoV-2 3CLpro substrate degradome. Cell Reports 37, 109892 (2021).
-
Gendrisch, F., Haarhaus, B., Schempp, C. M. & Wölfle, U. Anti-Psoriatic Effects of Antimony Compounds In Vitro. Molecules 26, 5814 (2021).
-
Daria Messelodi (2021). Study of pathway alterations in Gaucher disease by induced pluripotent stem cell models.
-
Stanfield, B.A. et al. (2021) IL-10 and class 1 histone deacetylases act synergistically and independently on the secretion of proinflammatory mediators in alveolar macrophages, PLOS ONE. Edited by P. Mukhopadhyay, 16(1), p. e0245169. doi:10.1371/journal.pone.0245169.
-
Licata, N.V. et al. (2021) C9orf72 ALS/FTD dipeptide repeat protein levels are reduced by small molecules that inhibit PKA or enhance protein degradation, The EMBO Journal [Preprint]. doi:10.15252/embj.2020105026.
-
Chandrasekaran, A. et al. (2021) Neural Derivates of Canine Induced Pluripotent Stem Cells-Like Cells From a Mild Cognitive Impairment Dog, Frontiers in Veterinary Science, 8, p. 725386. doi:10.3389/fvets.2021.725386.
-
Tidu, F. et al. (2021) NFAT signaling in human mesenchymal stromal cells affects extracellular matrix remodeling and antifungal immune responses, iScience, 24(6), p. 102683. doi:10.1016/j.isci.2021.102683.
-
Slavoff, S., Cao, X., Khitun, A., Harold, C., Bryant, C., Zheng, S.-J., Baserga, S., 2021. Nascent alt-protein chemoproteomics reveals a repressor of ribosome biogenesis (preprint). In Review. https://doi.org/10.21203/rs.3.rs-871945/v1
-
Tan, Y., and Song, J. (2021). Independent and Synergistic Modulations of Viscoelasticity and Stiffness of Dynamically Cross-Linked Cell-Encapsulating ClickGels by Covalently Tethered Polymer Brushes. Biomacromolecules 22, 3408–3415.
-
Lo Cascio, C., McNamara, J.B., Melendez, E.L., Lewis, E.M., Dufault, M.E., Sanai, N., Plaisier, C.L., and Mehta, S. (2021). Nonredundant, isoform-specific roles of HDAC1 in glioma stem cells. JCI Insight 6, e149232.
-
Castellanet, O., Ahmad, F., Vinik, Y., Mills, G.B., Habermann, B., Borg, J.-P., Lev, S., Lamballe, F., and Maina, F. (2021). BCL-XL blockage in TNBC models confers vulnerability to inhibition of specific cell cycle regulators. Theranostics 11, 9180–9197.
-
Arakaki, A.K.S., Pan, W.-A., Wedegaertner, H., Roca-Mercado, I., Chinn, L., Gujral, T.S., and Trejo, J. (2021). α-Arrestin ARRDC3 tumor suppressor function is linked to GPCR-induced TAZ activation and breast cancer metastasis. Journal of Cell Science 134, jcs254888.
-
Gendrisch, F., Haarhaus, B., Krieger, N., Quirin, K.-W., Schempp, C.M., and Wölfle, U. (2021). The Effect of Herbal Medicinal Products on Psoriasis-Like Keratinocytes. Biomolecules 11, 371.
-
Roggero, C. M. et al. Poly-glutamine-dependent self-association as a potential mechanism for regulation of androgen receptor activity. http://biorxiv.org/lookup/doi/10.1101/2021.10.08.463684 (2021) doi:10.1101/2021.10.08.463684.
-
Stanfield, B.A., Purves, T., Palmer, S., Sullenger, B., Welty-Wolf, K., Haines, K., Agarwal, S., and Kasotakis, G. (2021). IL-10 and class 1 histone deacetylases act synergistically and independently on the secretion of proinflammatory mediators in alveolar macrophages. PLoS ONE 16, e0245169.
-
Jiang, X., Maruyama, J., Iwasa, H., Arimoto-Matsuzaki, K., Nishina, H., and Hata, Y. (2021). Heat shock induces the nuclear accumulation of YAP1 via SRC. Experimental Cell Research 399, 112439.
-
Chen, L.-F., Lyons, M.R., Liu, F., Green, M.V., Hedrick, N.G., Williams, A.B., Narayanan, A., Yasuda, R., and West, A.E. (2020). The NMDA receptor subunit GluN3A regulates synaptic activity-induced and myocyte enhancer factor 2C (MEF2C)-dependent transcription. Journal of Biological Chemistry 295, 8613–8627.
-
Deville, S. S. et al. The Intermediate Filament Synemin Regulates Non-Homologous End Joining in an ATM-Dependent Manner. Cancers 12, 1717 (2020).
-
Caroline Stoten (2020). CHMP7 regulation during nuclear envelope reformation.
-
Victor Sebastian Tapia Olivare (2020). Understanding the mechanisms of Interleukin-1αprocessing and secretion. University of Manchester.
-
Fomicheva, M., and Macara, I.G. (2020). Genome-wide CRISPR screen identifies noncanonical NF-κB signaling as a regulator of density-dependent proliferation. ELife 9, e63603.
-
Panagiotakopoulou, V., Ivanyuk, D., De Cicco, S., Haq, W., Arsić, A., Yu, C., Messelodi, D., Oldrati, M., Schöndorf, D.C., Perez, M.-J., et al. (2020). Interferon-γ signaling synergizes with LRRK2 in neurons and microglia derived from human induced pluripotent stem cells. Nat Commun 11, 5163.
-
Deville, S.S., Delgadillo Silva, L.F., Vehlow, A., and Cordes, N. (2020). c-Abl Tyrosine Kinase Is Regulated Downstream of the Cytoskeletal Protein Synemin in Head and Neck Squamous Cell Carcinoma Radioresistance and DNA Repair. IJMS 21, 7277.
-
Xia, C., Wolf, J.J., Sun, C., Xu, M., Studstill, C.J., Chen, J., Ngo, H., Zhu, H., and Hahm, B. (2020). PARP1 Enhances Influenza A Virus Propagation by Facilitating Degradation of Host Type I Interferon Receptor. J Virol 94, e01572-19, /jvi/94/7/JVI.01572-19.atom.
-
Fisher, M.H., Kirkpatrick, G.D., Stevens, B., Jones, C., Callaghan, M., Rajpurkar, M., Fulbright, J., Cooper, M.A., Rowley, J., Porter, C.C., et al. (2020). ETV6 germline mutations cause HDAC3/NCOR2 mislocalization and upregulation of interferon response genes. JCI Insight 5, e140332.
-
Dill, T.L., Carroll, A., Gao, J., and Naya, F.J. (2020). The long noncoding RNA Meg3 regulates myoblast plasticity and muscle regeneration through epithelial-mesenchymal transition (Developmental Biology).
-
Guarnaccia, A.D., Rose, K.L., Wang, J., Zhao, B., Popay, T.M., Wang, C.E., Guerrazzi, K., Hill, S., Woodley, C.M., Hansen, T.J., et al. (2021). Impact of WIN site inhibitor on the WDR5 interactome. Cell Reports 34, 108636.
-
Lavau, C.P., Aumann, W.K., Sze, S.-G.K., Gupta, V., Ripple, K., Port, S.A., Kehlenbach, R.H., and Wechsler, D.S. (2020). The SQSTM1-NUP214 fusion protein interacts with Crm1, activates Hoxa and Meis1 genes, and drives leukemogenesis in mice. PLoS ONE 15, e0232036.
-
Wang, Y., Liu, Y., Nham, A., Sherbaf, A., Quach, D., Yahya, E., Ranburger, D., Bi, X., and Baudry, M. (2020). Calpain-2 as a therapeutic target in repeated concussion–induced neuropathy and behavioral impairment. Sci. Adv. 6, eaba5547.
-
Federico Tidu (2020). PRR SIGNALLING IN MESENCHYMAL STROMAL CELL RESPONSE TO FUNGAL INFECTION.
-
Endo, M., Tanaka, Y., Otsuka, M., and Minami, Y. (2020). E2F1‐Ror2 signaling mediates coordinated transcriptional regulation to promote G1/S phase transition in bFGF‐stimulated NIH/3T3 fibroblasts. FASEB J. 34, 3413–3428.
-
Lucchetti, D., Colella, F., Perelli, L., Ricciardi-Tenore, C., Calapà, F., Fiori, M.E., Carbone, F., De Maria, R., and Sgambato, A. (2020). CD147 Promotes Cell Small Extracellular Vesicles Release during Colon Cancer Stem Cells Differentiation and Triggers Cellular Changes in Recipient Cells. Cancers 12, 260.
-
Harman, J. (2019). Investigating the role of histone H3 lysine 9 dimethylation in regulating disease-associated vascular smooth muscle cell gene expression (Doctoral thesis). https://doi.org/10.17863/CAM.37204
-
Sociale, Mariangela: Ceramide Synthase in Transcriptional Regulation and Lipid Sensing. - Bonn, 2019. - Dissertation, Rheinische Friedrich-Wilhelms-Universität Bonn.
-
Zhong, X., Lee, H.-N., Kim, S.H., Park, S.-A., Kim, W., Cha, Y.-N., and Surh, Y.-J. (2018). Myc-nick promotes efferocytosis through M2 macrophage polarization during resolution of inflammation. The FASEB Journal 32, 5312–5325.
-
Wan, Y., and Hopper, A.K. (2018). From powerhouse to processing plant: conserved roles of mitochondrial outer membrane proteins in tRNA splicing. Genes & Development 32, 1309–1314.
-
Sociale, M., Wulf, A.-L., Breiden, B., Klee, K., Thielisch, M., Eckardt, F., Sellin, J., Bülow, M.H., Löbbert, S., Weinstock, N., et al. (2018). Ceramide Synthase Schlank Is a Transcriptional Regulator Adapting Gene Expression to Energy Requirements. Cell Reports 22, 967–978
-
Liang-Fu Chen (2017). Mechanisms of Specificity in Neuronal Activity-regulated Gene Transcription. Duke University.
-
Chulkina, M., Negmadjanov, U., Lebedeva, E., Pichugin, A., Mazurov, D., Ataullakhanov, R., and Holmuhamedov, E. (2017). Synthetic peptide TEKKRRETVEREKE derived from ezrin induces differentiation of NIH/3T3 fibroblasts. European Journal of Pharmacology 811, 249–259.
-
Purohit, M.P., Verma, N.K., Kar, A.K., Singh, A., Ghosh, D., and Patnaik, S. (2017). Inhibition of Thioredoxin Reductase by Targeted Selenopolymeric Nanocarriers Synergizes the Therapeutic Efficacy of Doxorubicin in MCF7 Human Breast Cancer Cells. ACS Applied Materials & Interfaces 9, 36493–36512.
-
Grbeša, I., Kalo, A., Belužić, R., Kovačević, L., Lepur, A., Rokić, F., Hochberg, H., Kanter, I., Simunović, V., Muńoz-Torres, P.M., et al. (2017). Mutations in S-adenosylhomocysteine hydrolase (AHCY) affect its nucleocytoplasmic distribution and capability to interact with S-adenosylhomocysteine hydrolase-like 1 protein. European Journal of Cell Biology 96, 579–590.
-
Sahoo, S.S., Quah, M.Y., Nielsen, S., Atkins, J., Au, G.G., Cairns, M.J., Nahar, P., Lombard, J.M., and Tanwar, P.S. (2017). Inhibition of extracellular matrix mediated TGF-β signalling suppresses endometrial cancer metastasis. Oncotarget.
-
Marina Chulkina, E.S. Lebedeva, Aleksei V Pichugin, E.L. Holmuhamedov, and Ravshan Inoyatovich Ataullakhanov (2017). Regeneration-inducing tetradecapeptide tekkrretvereke activates mapk-kinase signaling in NIH/3T3 fibroblasts. Immunologiya 38, 172–179.
-
Lucija Kovačević (2017). S-ADENOSYLHOMOCYSTEINE HYDROLASE DEFICIENCY: MOLECULAR MECHANISMS OF NOVEL DISORDER. University of Rijeka.
-
Lounis, M.A., Bergeron, K.-F., Burhans, M.S., Ntambi, J.M., and Mounier, C. (2017). Oleate activates SREBP-1 signaling activity in SCD1-deficient hepatocytes. American Journal of Physiology-Endocrinology and Metabolism 313, E710–E720.
-
Alexandra Candib (2016). Characterization and Validation of MGMT-binding Partners Using a Proteomic-based Approach in Glioblastoma. McGill University, Montreal, Canada
-
Dissertation zur Erlangung des Doktorgrades der Fakultät für Chemie und Pharmazie der Ludwig-Maximilians-Universität München. The Rabies Virus Phosphoprotein: Novel Targets and Functions Involved in Interferon Antagonism. Marco Wachowius (2016). Ludwig-Maximilians-University, Munich.