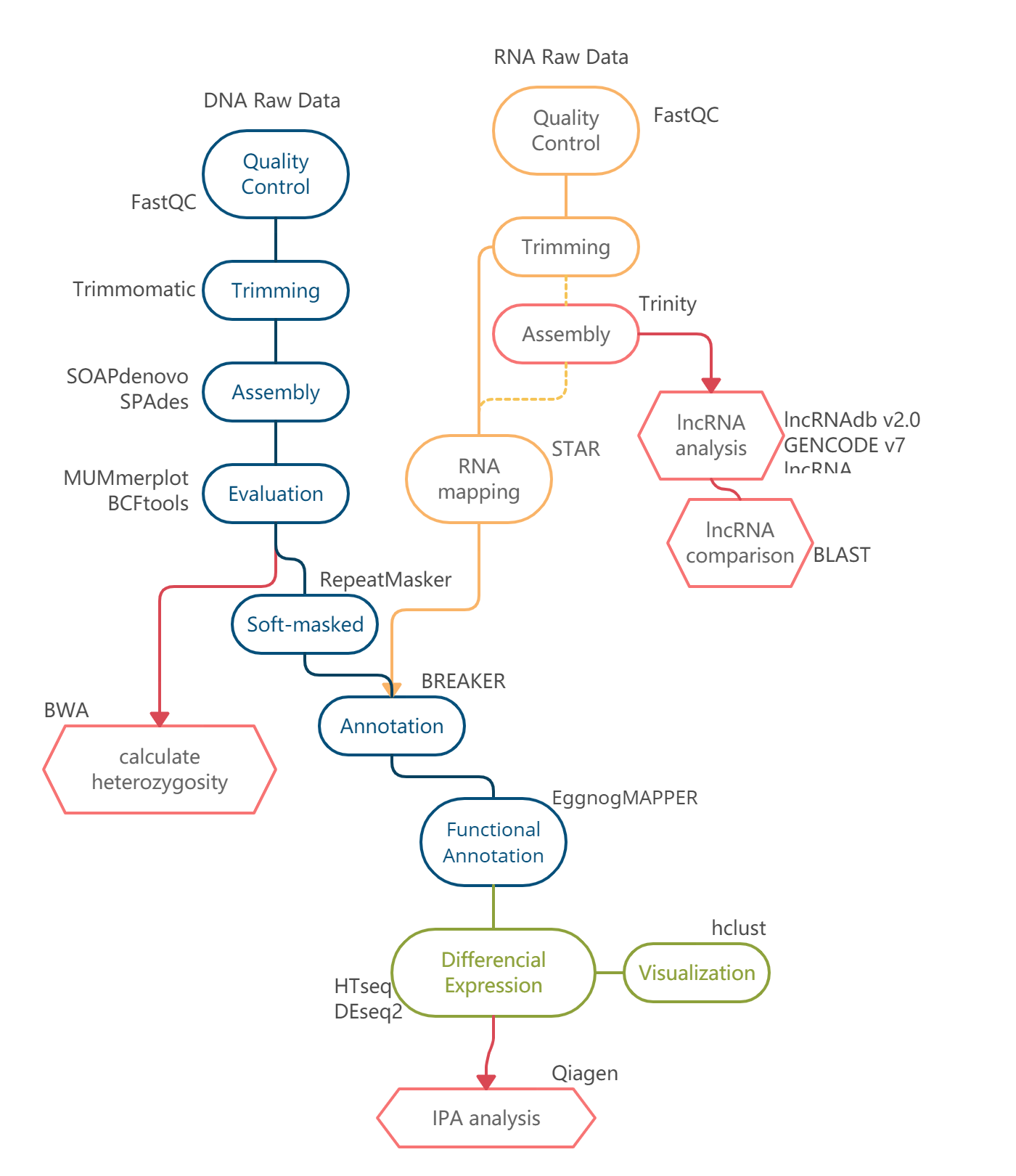
1. Overview - Kkkzq/Genome-Analysis-paper2 GitHub Wiki
1.1. Aim
The aim of this project is to detect genetic determinants related to the development of bat wings by characterizing Transcriptomic and epigenomic data. During the processes of this project, various bioinformatics tools were used and familiarized. It is necessary to mention that the results of this project might differ from the results in the paper because only part of the original data is used.
1.2. Time plan
| Day | Hours | Process | Status | Data Type | Software | Estimated Time | DDL |
|---|---|---|---|---|---|---|---|
| 26/3 | 2 | Seminar | Completed | -- | -- | 2h | Seminar |
| 1/4 | -- | Project Planning | Completed | -- | Github | 4h | -- |
| 6/4 | 4 | Reads quality control | Completed | Illumina | FastQC | 3min | Project planning |
| 6/4 | 4 | Reads preprocessing | Completed | Illumina | Trimmomatic | 1-10 min/file (2 cores) | Project planning |
| 9/4 | 4 | Genome Assembly | Completed | Illumina | SOAPdenovo | ~1.5h (2 cores) | -- |
| 9/4 | 4 | Assembly evaluation for SOAPdenovo (no gapcloser) | Completed | Fasta sequences | MUMmerplot | < 5min (1 core) | -- |
| 14/4 | 4 | Genome Assembly | Completed | Illumina | SPAdes | ~12 h (12 cores) | -- |
| 14/4 | 4 | Genome Assembly (after SOAPdenovo) | Completed | Illumina | GapCloser | -- | -- |
| 15/4 | 4 | Assembly evaluation for spades | Completed | Fasta sequences | MUMmerplot | < 5min (1 core) | Genome Assembly |
| 15/4 | 4 | Assembly evaluation for SOAPdenovo (after gapcloser) | Completed | Fasta sequences | MUMmerplot | < 5min (1 core) | -- |
| 15/4 | 4 | Transcriptome assembly | Incompleted | Illumina RNA | Trinity | ~1,5h (2 cores) | Genome Assembly |
| 20/4 | 4 | Genome Annotation | Completed | Eukaryotes | BRAKER | ? | -- |
| 20/4 | 4 | Functional Annotation | Completed | Eukaryotes | EggnogMAPPER | ? | -- |
| 28/4 | 4 | Calculate heterozygosity of genome | Incompleted | -- | BWA | ~1.5h (WGS,2cores) | -- |
| 29/4 | 4 | RNA mapping | Completed | Eukaryotes RNA | STAR | ? | Annotation |
| 29/4 | 4 | Differential Expression | Completed | -- | Htseq | < 5min 1 core | -- |
| 29/4 | 4 | Differential Expression | Completed | -- | DEseq2 | ? | -- |
| 3/5 | 4 | Compare lncRNAs to databases | Incompleted | lncRNA | lncRNAdb, GENCODE lncRNA | 1 h | -- |
| 3/5 | 4 | Compare homogenic lncRNAs | Incompleted | lncRNA | BLAST | 1 h | -- |
| 3/5 | 4 | Visualize ChIP-seq data | Incompleted | ChIP-seq data | DiffBind in R | ? | -- |
| 4/5 | 4 | Deeper analysis of differential expression | Partly Completed | -- | hclust, GO analysis | -- | RNA mapping |
| 17/5 | 4 | Check; finish wiki | Completed | -- | Github | -- | -- |
| 19/5 | 4 | Check; finish wiki | Completed | -- | Github | -- | -- |
| 24/5 | 4 | Final version of wiki | Incompleted | -- | Github | -- | Check all |
1.3. Data
DNA was extracted from the leg muscle tissue of a single male M. natalensis. RNA was extracted from paired forelimbs and hindlimbs from three individuals (biological replicates) at three developmental stages (CS15, CS16, and CS17). The subset of original data is provided through UPPMAX under /proj/g2021012/2_Eckalbar_2016.
There are 4 datasets which are sel1, sel2, sel3, and sel4 in this folder. Each dataset contains data from whole-genome sequencing, ChIP-sequencing, and RNA-sequencing. The additional folder contains the reference genome assemblies for each subset of data.
| Data | Tissue | Stage | Tool |
|---|---|---|---|
| WGS | Leg muscle of a single M. natalensis male | Adult | Illumina |
| ChIP-seq | Bat forelimb and hindlimb | CS15, CS16, CS17 | Illumina |
| RNA-seq | Bat forelimb and hindlimb | CS15, CS16, CS17 | Illumina |
WGS Data
The data in the following table is used for DNA assembly (SOAPdenovo).
| Read | Average insert Library size (bp) |
|---|---|
| SRR5819794 | 400 |
| SRR5819795 | 800 |
| SRR5819796 | 9000 |
| SRR5819797 | 175 |
| SRR5819798 | 3000 |
| SRR5819799 | 5500 |
RNA Data
| ID | Tissue | Development stage |
|---|---|---|
| SRR1719013 | Forelimb | CS15 |
| SRR1719014 | Forelimb | CS15 |
| SRR1719015 | Forelimb | CS15 |
| SRR1719016 | Hindlimb | CS15 |
| SRR1719017 | Hindlimb | CS15 |
| SRR1719018 | Hindlimb | CS15 |
| SRR1719204 | Forelimb | CS16 |
| SRR1719206 | Forelimb | CS16 |
| SRR1719207 | Forelimb | CS16 |
| SRR1719212 | Hindlimb | CS16 |
| SRR1719214 | Hindlimb | CS16 |
| SRR1719242 | Hindlimb | CS16 |
| SRR1719208 | Forelimb | CS17 |
| SRR1719209 | Forelimb | CS17 |
| SRR1719211 | Forelimb | CS17 |
| SRR1719213 | Hindlimb | CS17 |
| SRR1719241 | Hindlimb | CS17 |
| SRR1719266 | Hindlimb | CS17 |
These datasets including paired and unpaired reads.
Data Structure
The data and scripts of this project are stores in the following structure. The raw data is in the 2_Eckalbar_2016 and kept unchanged during the project. The unziped_data folder stores the data which has been copied and unzipped from the raw data folder. All scripts are in the script folder and all results are saved in the result folder.
.
├── 2_Eckalbar_2016 -> /proj/g2021012/2_Eckalbar_2016/
├── result
│ ├── 1_quality_control
│ ├── 2_trimming
│ ├── 3_dna_assembly
│ ├── 4_assembly_validation
│ ├── 5_transcriptome_assembly
│ ├── 6_repeatmasker
│ ├── 6_softmask_test
│ ├── 7_ref_mapping
│ ├── 7_rna_mapping
│ ├── 8_braker_annotation
│ ├── 8_ref_annotation
│ ├── 8_ref_eggnog
│ ├── 9_htseq
│ └── test
├── script
│ ├── 1_fastqc_chip.sh
│ ├── 1_fastqc_rna_raw_trimmed.sh
│ ├── 1_fastqc_rna.sh
│ ├── 1_fastqc_rna_trim.sh
│ ├── 1_fastqc_wgs.sh
│ ├── 2_trim_rna.sh
│ ├── 3_GapCloser_SOAP_wgs.sh
│ ├── 3_SOAPdenovo_wgs.sh
│ ├── 3_spades_assembly_wgs.sh
│ ├── 4_MUMmer_soapdenovo_nogapcloser_wgs.sh
│ ├── 4_MUMmer_soapdenovo_wgs.sh
│ ├── 4_MUMmer_spades_wgs.sh
│ ├── 5_trinity_rna_pair.sh
│ ├── 5_trinity_rna_single.sh
│ ├── 6_hard_repeatmasker.sh
│ ├── 6_repeatmasker_wgs.sh
│ ├── 6_repeatscout_wgs_filter.sh
│ ├── 6_repeatscout_wgs.sh
│ ├── 6_soft_repeatmasker.sh
│ ├── 7_ref_mapping.sh
│ ├── 7_rna_mapping.sh
│ ├── 7_test.sh
│ ├── 8_annotation_new.sh
│ ├── 8_BRAKER_annotation.sh
│ ├── 8_genemark.sh
│ ├── 8_ref_annotation.sh
│ ├── 8_ref_eggnog.sh
│ ├── 8_test.sh
│ ├── 9_htseq_count.sh
│ ├── slurm_log
│ ├── soapdenovo_config.txt
│ └── soapdenovo.log
└── unziped_data
├── GCF_001595765.1_Mnat.v1_genomic.gff
├── NW_015504249.1.gff
├── quast_output
├── sel2_NW_015504334.fna
└── sel3_NW_015504249.fna
21 directories, 38 files
1.4 Workflow