meeting 2023 08 01 n57 - JacobPilawa/TriaxSchwarzschild_wiki_5 GitHub Wiki
-
Some general information on N57:
- D = 66.9 ± 2.9 (Jensen+2021) – this is 10 Mpc different from the previous distance measurement of 76.3 Mpc
- GMOS PA: +41.0 E of N, Mitchell PA: +41.1;
- GMOS PSF: Using Avg. weighted FWHM = 0.81"; Mitchell PSF: 0.5 (from N1453+N2693)
- 215 GMOS bins, 41 Mitchell bins
- Also had done some preliminary sersic fits to determine R_core; could be interesting to revisit
-
First, here's a quick summary of our past cubes/models:
| Cube Name | Date | Summary |
|---|---|---|
| Cube A1 | Feb 3, 2022 | 6d hypercube, with gNFW profile. A bit undersampled, seemed to miss minimum. |
| Cube A2 | Feb 10, 2022 | Submitted 1000 more models to cover higher black hole and higher halo region. Points chosen to have ~uniform density in the space. |
| Cube A1A2 Scaled | Feb 22, 2022 | Scalings. Scaled the 13 models above to 13 different scales between 0.9 and 1.1, giving ~26,000 models total. |
| Cube B | March 1, 2022 | Rejection sampled cube incorrectly built from 26,000 scaled models. ~1000 rejection sampled models. The error was in applying the "scale" to the models. Also scaled these models. |
| Cube B Prime | March 15, 2022 | Corrected the scaling issue from Cube B and ran ~1000 more models. Also scaled these models. |
| Cube C | March 21, 2022 | Rejection cube of 1000 models built from Cube A1A2 + Cube B Prime. |
| Lipka Tests | March-April 2022 | 729 models at fixed masses, testing any effect of the Lipka m_eff on the shapes for N57. |
| Cube D | May 17, 2022 | Built another rejection sampled cube based on the 1sigma contours (instead of 2 sigma contours) to try to solve the issue bi-modality issue in Tmaj. Didn't seem to improve things that much. |
Chi2 vs. Parameters for EVERY Model + Contours
 |
 |
 |
 |
 |
 |
 |
 |
Plots of the various cubes in chi2 vs. parameter space
CubeA1A2
| A1 | A2 |
|---|---|
 |
 |
Cube B + B'
| B | B' |
|---|---|
 |
 |
Cube C
| C |
|---|
 |
Cube D
| D |
|---|
 |
Lipka Test
| Lipka |
|---|
 |
A Scalings
| A1 | 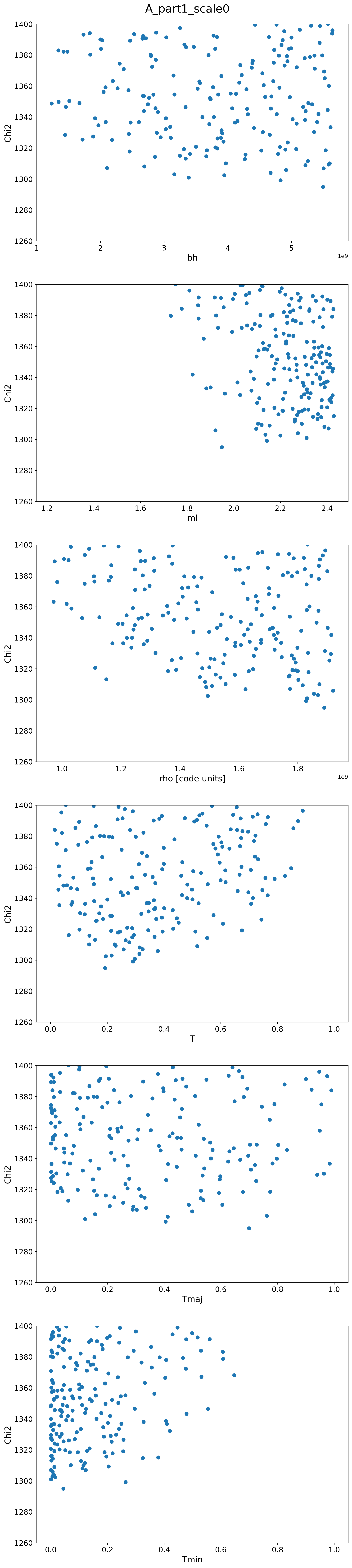 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| A2 |  |
 |
 |
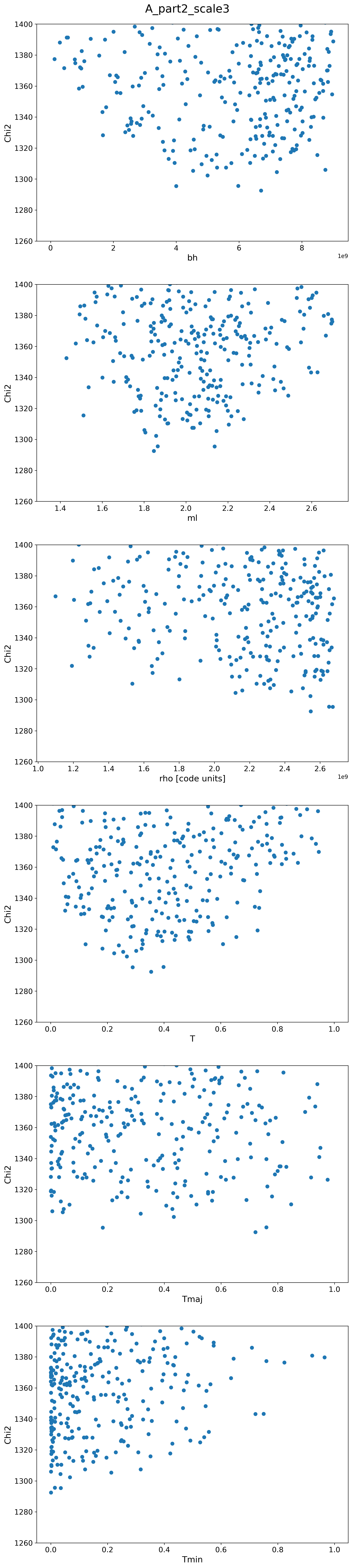 |
 |
 |
 |
 |
 |
 |
 |
 |
B Scalings
| B |  |
 |
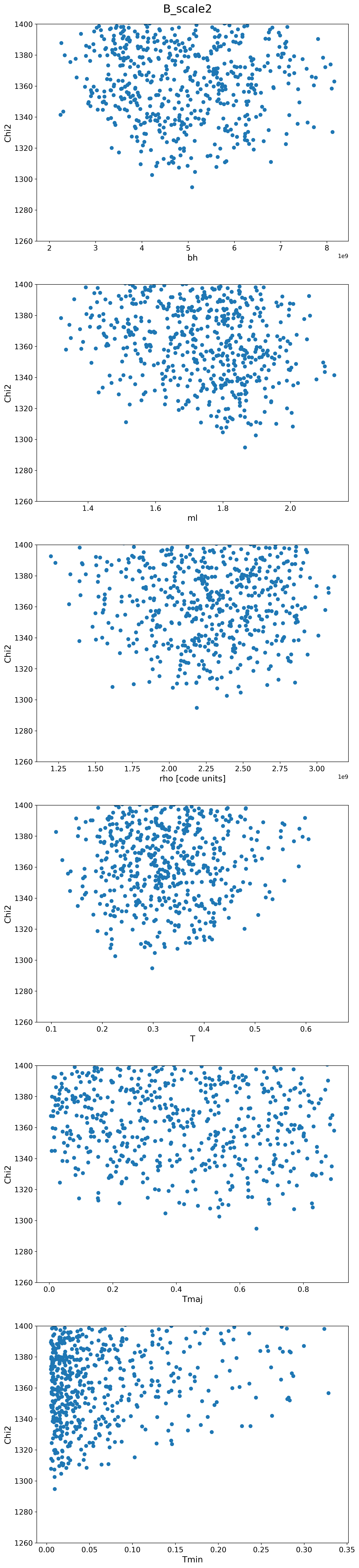 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| B' |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
All Fiducials
 |
All Fiducials No Lipka
 |
Here's an old, naive version of the cornerplot where I simply throw all models at GPR + dynesty with a cutoff K = 30. This assumed a chi2 error of 0.5 which is what we have used in the past.
| niter=1 | niter=2 | niter=4 | |
|---|---|---|---|
| Chi2 error = 0.5 |  |
 |
 |
And here are the diagnostics of the best fitting model so far
| Input 1 |  |
| Input 2 |  |
| Input 3 |  |
| Input 4 |  |
| Input 5 |  |
| Input 6 |  |
| Profiles 1 |  |
| Profiles 2 |  |
| Profiles 3 |  |
| Profiles 4 |  |
| Profiles 5 |  |
| Profiles 6 |  |
| Beta |  |
| Betaz |  |
| Menc |  |
And lastly, here are the diagnostics broken down by bin and by moment (I quite like this plot!) to see if there are any obvious trends or features in our data driving our chi2 landscapes.
 |
 |
|
 |
- Last time we were looking in detail at N57, we were playing around with a thinning routine to see if the "islands" we saw in our posteriors were real. We have done a good deal of work since then so I don't think this is necessarily the best approach, but I wanted to summarize what we have already done here:
- I thinned the full set of points into 20 realizations of ~1200 model points, each containing ~150 points within the 1 sigma contours
- It seemed like the 20 realizations all agreed quite well with one another, and combining these into a single chain produces a "nice looking" set of posteriors
- I thinned the full set of points into 20 realizations of ~1200 model points, each containing ~150 points within the 1 sigma contours
Here's quick plots showing the number of models vs. confidence interval/Cutoff K value
| vs. Delta Chi2 | vs. Confidence Level | |
|---|---|---|
 |
 |
Here's a single cornerplot with all the realizations plotted on top of each other
 |
For completeness, here are all the cornerplots but plotted separately so that we can see the individual landscapes.
| r1 | r2 | r3 | r4 | r5 | r6 | r7 | r8 | r9 | r10 | r11 | r12 | r13 | r14 | r15 | r16 | r17 | r18 | r19 | r20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
Lastly, here's the results of combining all samples into a single chain and plotting the contours
 |
 |
-
Emily's selection procedure for the scaled cases
- We discussed last time that taking the best-performing chi2 from each mass model (instead of all the scaled versions) is likely a better approach than anything we have done so far, so I can go ahead and implement that.
-
Specific checks to see about sampling in T/Tmaj/Tmin or (u,p,q) as we have been discussing