Requirements for opening a Nursery or Trial in the BMS - IntegratedBreedingPlatform/Documentation GitHub Wiki
This document is intended to support Data Managers in setting data correctly in the database for the BMS to access in the Nursery and Trial Managers
Opening an Existing Nursery
The Nursery and Trial records in the BMS reside in the project table as a collection of datasets. By default, when a Study is created, 3 datasets are created
- Study Details (the main dataset)
- Trial Dataset (also referred to as Summary Dataset and Trial Environment Dataset)
- Plotdata Dataset (also referred to as Measurement Dataset)
These records should have the program UUID that refers to the program they belong to, found in the workbench_project.program_uuid field. The program_uuid field can be the following :
| The program_uuid cross referenced to workbench_project.program_uuid | Behaviour |
|---|---|
| Valid UUID string | Appears in Folder Tree, is accessible as editable |
| Empty string or incorrect string | Will not appear in Folder Tree |
| NULL | Appears in Folder Tree, opens as Read-Only |
Opening a Nursery/Trial as 3 Datasets
Each dataset is most efficiently identified by a suffix to the Dataset name. The system looks for :
- Study Dataset : identified always by Id
- Trial Dataset : suffix "–ENVIRONMENT"
- Measurement Dataset : suffix "-PLOTDATA"
If these suffices are not found, then the BMS will travel to DB to fetch the Dataset Type, and will look for the following values
- Trial Dataset : cvterm_id = 10080
- Measurement Dataset : cvterm_id = 10090
*** means dataset is also recognised as 10070, but not in the opening of a Nursery/Trial for edit
If possible, using the name identification successfully will improve performance. The system is, however OK to search for dataset type in the projectprop table.
If neither of these methods are successful, then the BMS will return an exception to the user.
How Datasets Are Created
(we can only imagine this is what the Software expects in return)
The datasets are essentially group of variables prepared in such a way for analysis
You can see the screenshot below describes which variables get stored in the DB and which dataset these variables are housed.
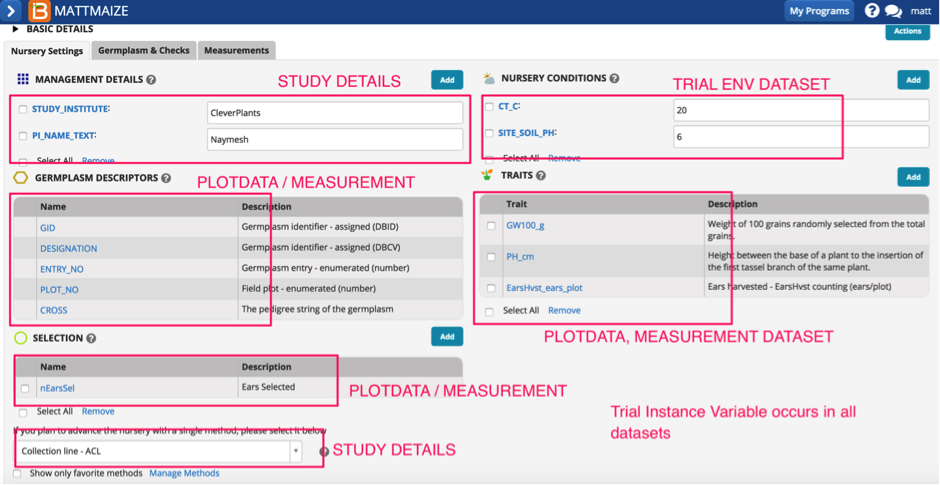
*** note and apologies that CT_C is noted here as a Condition, when it is in fact a Trait. Example only.
Here we have examples of all three datasets that the Nursery Manager stores – you will be able to see the added variables and the default variables in the dataset defined by the projectprop table for a particular project.
Example Datasets
Projects

Study (basic) Dataset
See in highlight that values for this dataset are stored directly in the projectprop table
| projectprop_id | project_id | type_id | value | rank |
|---|---|---|---|---|
| 26197 | 25035 | 1060 | Study type - assigned (type) | 1 |
| 26198 | 25035 | 1070 | 8070 | 1 |
| 26199 | 25035 | 8070 | 10000 | 1 |
| 26196 | 25035 | 1805 | STUDY_TYPE | 1 |
| 26166 | 25035 | 1805 | STUDY_BM_CODE | 2 |
| 26168 | 25035 | 1070 | 8251 | 2 |
| 26167 | 25035 | 1060 | Breeding method applied to all plots in a study (CODE) | 2 |
| 26200 | 25035 | 8251 | ACL | 2 |
| 26170 | 25035 | 1060 | Study - assigned (DBCV) | 3 |
| 26169 | 25035 | 1805 | STUDY_NAME | 3 |
| 26201 | 25035 | 8005 | HandsonTableTest | 3 |
| 26171 | 25035 | 1070 | 8005 | 3 |
| 26172 | 25035 | 1805 | STUDY_TITLE | 4 |
| 26174 | 25035 | 1070 | 8007 | 4 |
| 26173 | 25035 | 1060 | Study title - assigned (text) | 4 |
| 26202 | 25035 | 8007 | Testing JS table | 4 |
| 26203 | 25035 | 8050 | 20151031 | 5 |
| 26177 | 25035 | 1070 | 8050 | 5 |
| 26176 | 25035 | 1060 | Start date - assigned (date) | 5 |
| 26175 | 25035 | 1805 | START_DATE | 5 |
| 26204 | 25035 | 8030 | Testing The Objective | 6 |
| 26179 | 25035 | 1060 | Objective - described (text) | 6 |
| 26178 | 25035 | 1805 | STUDY_OBJECTIVE | 6 |
| 26180 | 25035 | 1070 | 8030 | 6 |
| 26205 | 25035 | 8060 | 20160301 | 7 |
| 26181 | 25035 | 1805 | END_DATE | 7 |
| 26183 | 25035 | 1070 | 8060 | 7 |
| 26182 | 25035 | 1060 | End date - assigned (date) | 7 |
| 26206 | 25035 | 8020 | 52 | 8 |
| 26184 | 25035 | 1805 | Study_UID | 8 |
| 26186 | 25035 | 1070 | 8020 | 8 |
| 26185 | 25035 | 1060 | ID of the user entering the study data - assigned (DBID) | 8 |
| 26188 | 25035 | 1060 | Date the study was last updated - assigned (YYYYMMDD) | 9 |
| 26189 | 25035 | 1070 | 8009 | 9 |
| 26187 | 25035 | 1805 | STUDY_UPDATE | 9 |
| 26207 | 25035 | 8009 | 20151104 | 9 |
| 26192 | 25035 | 1070 | 8256 | 10 |
| 26190 | 25035 | 1805 | STUDY_BMETH | 10 |
| 26208 | 25035 | 8256 | Collection line | 10 |
| 26191 | 25035 | 1060 | Breeding method applied to all plots (DBCV) | 10 |
| 26194 | 25035 | 1060 | TRIAL_INSTANCE | 11 |
| 26195 | 25035 | 1070 | 8170 | 11 |
| 26193 | 25035 | 1806 | TRIAL_INSTANCE | 11 |
| 26396 | 25035 | 1070 | 8080 | 12 |
| 26397 | 25035 | 8080 | CleverPlants | 12 |
| 26395 | 25035 | 1060 | Study institute - conducted (DBCV) | 12 |
| 26394 | 25035 | 1805 | STUDY_INSTITUTE | 12 |
| 26400 | 25035 | 1070 | 8115 | 13 |
| 26401 | 25035 | 8115 | Naymesh | 13 |
| 26398 | 25035 | 1805 | PI_NAME_TEXT | 13 |
| 26399 | 25035 | 1060 | Principal investigator - assigned (Name) | 13 |
Trial (environment) Dataset
Values for this dataset are stored in the geolocationprop table
*** note and apologies that CT_C is noted here as a Condition, when it is in fact a Trait. Example only
| projectprop_id | project_id | type_id | value | rank |
|---|---|---|---|---|
| 26216 | 25036 | 1060 | Dataset name | 1 |
| 26217 | 25036 | 1070 | 8150 | 1 |
| 26215 | 25036 | 1805 | DATASET_NAME | 1 |
| 26219 | 25036 | 1060 | Dataset title | 2 |
| 26220 | 25036 | 1070 | 8155 | 2 |
| 26218 | 25036 | 1805 | DATASET_TITLE | 2 |
| 26222 | 25036 | 1060 | Dataset type | 3 |
| 26223 | 25036 | 1070 | 8160 | 3 |
| 26221 | 25036 | 1805 | DATASET_TYPE | 3 |
| 26224 | 25036 | 8160 | 10080 | 3 |
| 26210 | 25036 | 1060 | Experimental design - assigned (type) | 4 |
| 26211 | 25036 | 1070 | 8135 | 4 |
| 26209 | 25036 | 1806 | EXPT_DESIGN | 4 |
| 26213 | 25036 | 1060 | TRIAL_INSTANCE | 5 |
| 26214 | 25036 | 1070 | 8170 | 5 |
| 26212 | 25036 | 1806 | TRIAL_INSTANCE | 5 |
| 26406 | 25036 | 1060 | Measured in several rows simultaneously to allow a comparison. Thermal images will be taken using an Infrared Camera placed several meters above the canopy. | 7 |
| 26407 | 25036 | 1070 | 20397 | 7 |
| 26405 | 25036 | 1808 | CT_C | 7 |
| 26409 | 25036 | 1060 | Soil acidity - ph meter (pH) | 8 |
| 26410 | 25036 | 1070 | 8270 | 8 |
| 26408 | 25036 | 1808 | SITE_SOIL_PH | 8 |
Measurement (plotdata) Dataset
| projectprop_id | project_id | type_id | value | rank |
|---|---|---|---|---|
| 26244 | 25037 | 1060 | Dataset name | 1 |
| 26245 | 25037 | 1070 | 8150 | 1 |
| 26243 | 25037 | 1805 | DATASET_NAME | 1 |
| 26247 | 25037 | 1060 | Dataset title | 2 |
| 26248 | 25037 | 1070 | 8155 | 2 |
| 26246 | 25037 | 1805 | DATASET_TITLE | 2 |
| 26250 | 25037 | 1060 | Dataset type | 3 |
| 26251 | 25037 | 1070 | 8160 | 3 |
| 26249 | 25037 | 1805 | DATASET_TYPE | 3 |
| 26252 | 25037 | 8160 | 10090 | 3 |
| 26226 | 25037 | 1060 | TRIAL_INSTANCE | 4 |
| 26227 | 25037 | 1070 | 8170 | 4 |
| 26225 | 25037 | 1806 | TRIAL_INSTANCE | 4 |
| 26229 | 25037 | 1060 | Germplasm identifier - assigned (DBID) | 5 |
| 26230 | 25037 | 1070 | 8240 | 5 |
| 26228 | 25037 | 1804 | GID | 5 |
| 26232 | 25037 | 1060 | Germplasm identifier - assigned (DBCV) | 6 |
| 26233 | 25037 | 1070 | 8250 | 6 |
| 26231 | 25037 | 1804 | DESIGNATION | 6 |
| 26235 | 25037 | 1060 | Germplasm entry - enumerated (number) | 7 |
| 26236 | 25037 | 1070 | 8230 | 7 |
| 26234 | 25037 | 1804 | ENTRY_NO | 7 |
| 26238 | 25037 | 1060 | Field plot - enumerated (number) | 8 |
| 26239 | 25037 | 1070 | 8200 | 8 |
| 26237 | 25037 | 1810 | PLOT_NO | 8 |
| 26241 | 25037 | 1060 | The pedigree string of the germplasm | 9 |
| 26242 | 25037 | 1070 | 8377 | 9 |
| 26240 | 25037 | 1804 | CROSS | 9 |
| 26254 | 25037 | 1060 | Weight of 100 grains randomly selected from the total grains. | 10 |
| 26255 | 25037 | 1070 | 51496 | 10 |
| 26253 | 25037 | 1808 | GW100_g | 10 |
| 26257 | 25037 | 1060 | Height between the base of a plant to the insertion of the first tassel branch of the same plant. | 11 |
| 26258 | 25037 | 1070 | 20343 | 11 |
| 26256 | 25037 | 1808 | PH_cm | 11 |
| 26260 | 25037 | 1060 | Ears harvested - EarsHvst counting (ears/plot) | 12 |
| 26261 | 25037 | 1070 | 51497 | 12 |
| 26259 | 25037 | 1808 | EarsHvst_ears_plot | 12 |
| 26412 | 25037 | 1060 | Ears Selected | 13 |
| 26413 | 25037 | 1070 | 20364 | 13 |
| 26411 | 25037 | 1808 | nEarsSel | 13 |
Open Study
- getStudyDetails – main dataset for a Nursery/Trial (contains trial and measurement dataset ids)
- get trialDataSet – get Study by Id
- get measurementDataset – get Study by id
- getExperiments – from experiment_project
- getTrial Instances – from geolocation
- getLocationInfo – from geolocationprop
- BreedingMethods – non generative
Study
This is the main query to fetch Study information – you can see the tables we are joining to get this information. This query returns data for the main "Basic Details" section of the Nursery and Trial Manager. You will see we hit Locations and experiments as well but this information is not used and should be trimmed from the query.
StringBuilder sqlString = **new** StringBuilder()
`.append("SELECT DISTINCT p.name AS name, p.description AS title, ppObjective.value AS objective, ppStartDate.value AS startDate,`
ppEndDate.value AS endDate, ppPI.value AS piName, gpSiteName.value AS siteName, p.project\_id AS id, ppPIid.value AS piId, gpSiteId.value AS siteId, ppFolder.object\_project\_id AS folderId, p.program\_uuid AS programUUID FROM project p ")
INNER JOIN projectprop ppNursery ON p.project\_id = ppNursery.project\_id
AND ppNursery.type\_id = ")
`.append(TermId. **STUDY\_TYPE**.getId())`
`.append(" ")`
`.append("AND ppNursery.value = ")`
`.append(studyType.getId())`
`.append(" ")`
`.append("INNER JOIN project\_relationship ppFolder ON p.project\_id = ppFolder.subject\_project\_id ")`
`.append(" LEFT JOIN projectprop ppObjective ON p.project\_id = ppObjective.project\_id ")`
`.append("AND ppObjective.type\_id = ")`
`.append(TermId. **STUDY\_OBJECTIVE**.getId())`
`.append(" ")`
`.append(" LEFT JOIN projectprop ppStartDate ON p.project\_id = ppStartDate.project\_id ")`
`.append("AND ppStartDate.type\_id = ")`
`.append(TermId. **START\_DATE**.getId())`
`.append(" ")`
`.append(" LEFT JOIN projectprop ppEndDate ON p.project\_id = ppEndDate.project\_id ")`
.append(" AND ppEndDate.type\_id = ")
`.append(TermId. **END\_DATE**.getId())`
`.append(" ")`
`.append(" LEFT JOIN projectprop ppPI ON p.project\_id = ppPI.project\_id ") .append(" AND ppPI.type\_id = ")`
`.append(TermId. **PI\_NAME**.getId())`
`.append(" ")`
`.append(" LEFT JOIN projectprop ppPIid ON p.project\_id = ppPIid.project\_id ")`
`.append(" AND ppPIid.type\_id = ")`
`.append(TermId. **PI\_ID**.getId())`
`.append(" ")`
`.append(" LEFT JOIN nd\_experiment\_project ep ON p.project\_id = ep.project\_id ")`
`.append(" LEFT JOIN nd\_experiment e ON ep.nd\_experiment\_id = e.nd\_experiment\_id ")`
`.append(" LEFT JOIN nd\_geolocationprop gpSiteName ON e.nd\_geolocation\_id = gpSiteName.nd\_geolocation\_id ")`
.append(" AND gpSiteName.type\_id = ").append(TermId. **TRIAL\_LOCATION**.getId()).append(" ")
`.append(" LEFT JOIN nd\_geolocationprop gpSiteId ON e.nd\_geolocation\_id = gpSiteId.nd\_geolocation\_id ")`
`.append(" AND gpSiteId.type\_id = ").append(TermId. **LOCATION\_ID**.getId()).append(" ")`
`.append(" WHERE p.project\_id = ").append(studyId);`
Trial Dataset
This is the dataset that holds the Managerial and Environmental Conditions for a Nursery. We pick this up from the database using our generic 'getDataset' machinery, and passing the dataset ID. It is essentially the same as fetching any study from the DB.
Hibernate:
`/\* load org.generationcp.middleware.pojos.dms.DmsProject \*/ select`
`dmsproject0\_.project\_id as project1\_852\_0\_,`
`dmsproject0\_.description as descript2\_852\_0\_,`
`dmsproject0\_.name as name852\_0\_,`
`dmsproject0\_.program\_uuid as program4\_852\_0\_`
`from`
`project dmsproject0\_`
`where`
`dmsproject0\_.project\_id=?`
Hibernate:
`/\* load one-to-many org.generationcp.middleware.pojos.dms.DmsProject.relatedTos \*/ select`
`relatedtos0\_.subject\_project\_id as subject4\_852\_2\_,`
`relatedtos0\_.project\_relationship\_id as project1\_2\_,`
`relatedtos0\_.project\_relationship\_id as project1\_862\_1\_,`
`relatedtos0\_.object\_project\_id as object3\_862\_1\_,`
`relatedtos0\_.subject\_project\_id as subject4\_862\_1\_,`
`relatedtos0\_.type\_id as type2\_862\_1\_,`
`dmsproject1\_.project\_id as project1\_852\_0\_,`
`dmsproject1\_.description as descript2\_852\_0\_,`
`dmsproject1\_.name as name852\_0\_,`
`dmsproject1\_.program\_uuid as program4\_852\_0\_`
`___from___`
`___project\_relationship relatedtos0\____`
`___inner join___`
`___project dmsproject1\____`
`___on relatedtos0\_.object\_project\_id=dmsproject1\_.project\_id___`
`___where___`
`___relatedtos0\_.subject\_project\_id=?___`
Hibernate:
`/\* load one-to-many org.generationcp.middleware.pojos.dms.DmsProject.properties \*/ select`
`properties0\_.project\_id as project5\_852\_1\_,`
`properties0\_.projectprop\_id as projectp1\_1\_,`
`properties0\_.projectprop\_id as projectp1\_853\_0\_,`
`properties0\_.project\_id as project5\_853\_0\_,`
`properties0\_.rank as rank853\_0\_,`
`properties0\_.type\_id as type3\_853\_0\_,`
`properties0\_.value as value853\_0\_`
`from`
`projectprop properties0\_`
`where`
`properties0\_.project\_id=?`
Hibernate:
`/\* dynamic native SQL query \*/ SELECT`
`DISTINCT e.nd\_geolocation\_id`
`FROM`
`nd\_experiment e,`
`nd\_experiment\_project ep`
`WHERE`
`e.nd\_experiment\_id = ep.nd\_experiment\_id`
`and ep.project\_id = 25036`
Experiments (Measurements)
StringBuilder queryString = **new** StringBuilder();
queryString.append("select distinct ep from ExperimentProject as ep ");
queryString.append("inner join ep.experiment as exp ");
queryString.append("left outer join exp.properties as plot with plot.typeId IN (8200,8380) ");
queryString.append("left outer join exp.properties as rep with rep.typeId = 8210 ");
queryString.append("left outer join exp.experimentStocks as es ");
queryString.append("left outer join es.stock as st ");
queryString.append("where ep.projectId =:p\_id and ep.experiment.typeId in (:type\_ids) ");
queryString.append("order by (ep.experiment.geoLocation.description \* 1) ASC, ");
queryString.append("(plot.value \* 1) ASC, ");
queryString.append("(rep.value \* 1) ASC, ");
queryString.append("(st.uniqueName \* 1) ASC, ");
queryString.append("ep.experiment.ndExperimentId ASC");
Trial Instances for a study
String sql =
"SELECT DISTINCT e.nd\_geolocation\_id " + " FROM nd\_experiment e "
+ " INNER JOIN nd\_experiment\_project ep ON ep.nd\_experiment\_id = e.nd\_experiment\_id "
+ " INNER JOIN project\_relationship pr ON pr.type\_id = " + TermId. **BELONGS\_TO\_STUDY**.getId()
+ " AND pr.object\_project\_id = " + studyId + " AND pr.subject\_project\_id = ep.project\_id ";
Assembly
Once all these data are fetched, the BMS builds the following and assembles it into a data structure that carries data to the screen
workbook.setStudyDetails(studyDetails); - study **details** query
workbook.setFactors(factors); - from whole study FOO
workbook.setVariates(variates); - from whole study FOO
workbook.setConditions(conditions); - from whole Study
workbook.setConstants(constants); - from whole study
workbook.setObservations(observations); - from
workbook.setTreatmentFactors(treatmentFactors); - from
workbook.setExperimentalDesignVariables(expDesignVariables); - from Experiment Query
workbook.setTrialObservations(trialObservations); - from Experiment Query