2016 Key Workflows Analysis (Historic) - IntegratedBreedingPlatform/Documentation GitHub Wiki
Create Trial
Profiling observations
Defining Mega Trial as trial with 50 Germplasm entries, 3 replications, 25 Traits, 200 Locations with each location having one location property LOCATION_ABBR and one condition SITE_SOIL_PH. No measurements as we can not create measurements before first save.
There are two major areas where most of the backend time is spent:
- Experimental design: Takes an average of 38,552 ms for generating randomized complete block design for 200 environments. BMS spawns VSNI BVDesign engine Windows executable in loop for each environment which is indeed time consuming. Design per location might be a limitation of the VSNI engine, we are not entirely certain.
- Insert Trial Data Records: Average time spent at the backend for saving trial data to database, was 26,943 ms. Most of the time is spent in inserts related to observation units (
nd_experimenttable) and their design related properties (nd_experimentproptable). See the query stats below for details.
Query Stats
Core query stats for creating mega trial:
- Number of selects : 215,720 (214,026 of them are: select @@session.tx_read_only. Eliminated by adding useLocalSessionState=true in MySQL driver connection properties). So 1694 selects from BMS. 722 prepare select statements, 1448 execute selects.
- Number of inserts : 151,934 (includes prepare statements)
Key categories of database inserts:
-
Create trial environment/instance/location (nd_geolocation/nd_geolocationprop) records. 4 inserts per environment as below. Total 800 inserts.
insert into nd_geolocation (altitude, description, geodetic_datum, latitude, longitude) values (NULL, '1', NULL, NULL, NULL)
insert into nd_geolocationprop (nd_geolocation_id, rank, type_id, value) values (1, 2, 8189, 'Loc1') <-- 8189 = LOCATION_ABBR insert into nd_geolocationprop (nd_geolocation_id, rank, type_id, value) values (1, 3, 8135, '10110') <-- 8135 = EXPT_DESIGN insert into nd_geolocationprop (nd_geolocation_id, rank, type_id, value) values (1, 4, 8131, '3') <-- 8131 = NREP
-
Create Basic trial information (project/projectprop) records.
3 inserts into project (3 datasets) 161 inserts into projectprops (We only have one extra attribute STUDY_INSTITUTE = CIMMYT in settings tab)
3 inserts (cvterm id, name, definition definintion) for property name and 1 insert for property value, e.g. study name is stored with:
insert into projectprop (project_id, rank, type_id, value) values (25007, 2, 1805, 'STUDY_NAME') <-- 1805 = Study Detail insert into projectprop (project_id, rank, type_id, value) values (25007, 2, 1060, 'Study - assigned (DBCV)') <-- 1060 = Variable description insert into projectprop (project_id, rank, type_id, value) values (25007, 2, 1070, '8005') <-- 1070 = Standardvariableid insert into projectprop (project_id, rank, type_id, value) values (25007, 2, 8005, 'Mega Trial 764') <-- 8005 = STUDY_NAME -
Create observation unit (plots) and related information records. MAJOR HOTSPOT
200 inserts for the SITE_SOIL_PH value for each environment condition insert into phenotype (assay_id, attr_id, cvalue_id, name, observable_id, uniquename, value, phenotype_id) values (NULL, NULL, NULL, '8270', 8270, NULL, '6', 500) <-- 8270 = SITE_SOIL_PH
200 inserts for join table for the SITE_SOIL_PH value for each environment condition insert into nd_experiment_phenotype (nd_experiment_id, phenotype_id, nd_experiment_phenotype_id) values (501, 500, 500)
30000 inserts into nd_experiment 30000 inserts into nd_experiment_project (join table) 60000 inserts into nd_experimentprop table (because of two properties per plot PLOT_NO and REP_NO)
insert into nd_experimentprop (nd_experiment_id, rank, type_id, value) values (702, 9, 8210, '1') <-- 8210 = REP_NO insert into nd_experimentprop (nd_experiment_id, rank, type_id, value) values (702, 10, 8200, '2') <-- 8200 = PLOT_NO30000 inserts into nd_experiment_stock (join table)
insert into nd_experiment_stock (nd_experiment_id, stock_id, type_id) values (701, 1, 1000) <-- 1000= IBDB structure. Dont know why we do this! insert into nd_experiment_stock (nd_experiment_id, stock_id, type_id) values (702, 2, 1000) insert into nd_experiment_stock (nd_experiment_id, stock_id, type_id) values (703, 3, 1000) ... insert into nd_experiment_stock (nd_experiment_id, stock_id, type_id) values (30700, 21, 1000)50 inserts into stock (50 entries) 50 inserts into stockprop (50 entries entry type value for each)
insert into stockprop (rank, stock_id, type_id, value) values (5, 1, 8255, '10170') <-- 8255 = ENTRY_TYPE, 10170 = Test Entry -
Save snapshot of germplasm list for trial (ListData_Project - list copy)
1 insert into listnms for saving list copy name insert into listnms (listdate, listdesc, eDate, listlocn, listref, listname, notes, lhierarchy, program_uuid, projectId, sdate, liststatus, listtype, listuid) values (20160616, 'Test of large germplasm list import', NULL, NULL, 2, '50 maize list test', '', NULL, '36ad2439-9e16-47ae-bf72-355be4c1a764', 25007, NULL, 1, 'TRIAL', 1)
50 inserts into listdata_project to save list entries like:
insert into listdata_project (check_type, designation, duplicate_notes, entry_code, entry_id, germplasm_id, group_name, list_id, seed_source) values (10170, 'CML502-123', NULL, '1', 1, 5, 'CML176/CML264', 3, 'AF06A-251-2')
JAMon Timing markers
- On Windows environment (Quad Core 16GB RAM on Google Cloud) with VSNi BVDesign engine:
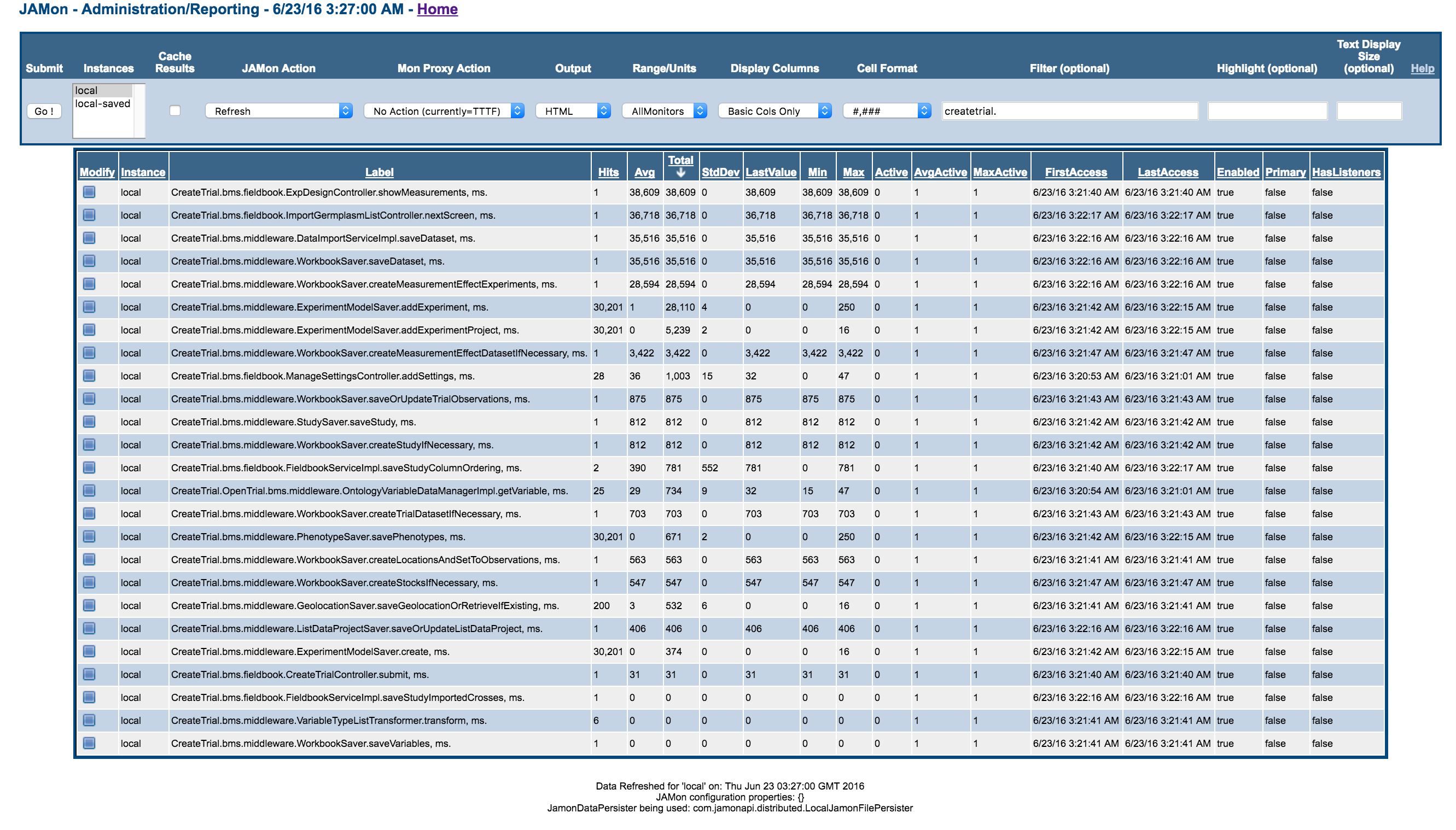
- On Mac OSX (2.4 GHz, 8GB RAM) with mocked design engine (hence the design generation part shows low timings)

Open Trial
Query stats:
- Total select queries executed: 1241. Of these 527 are preparing the select statements and 617 of them are actual execution of selects. 43 other queries e.g connection pool test queries (Select 1).
- Of the 617 selects:
- 68 selects from
nd_experimentbatched for 500 rows at a time to load 30000 observation units. - 65 selects from
nd_experimentproptable to load the experiment properties (design coordinates). Batched for 500 experiments per query. There are a total of 60000 props for our mega trial. - 65 selects from
nd_experiment_phenotype(join table to get to plot's phenotypes) batch of 500 phenotype units at a time. - 65 selects from
nd_experiment_stock(join table to get plot's germplasm info) batch of 500 as well. There are a total of 30000 records in this table for our mega trial. - 200 selects from
nd_geolocationprop- this can be easily reduced by adding batch loading. - 88 selects to
cvtermand related tables. - Other minor queries for stock, stockprop, project, projectprop, user etc. tables
- 68 selects from
JAMon Timing markers
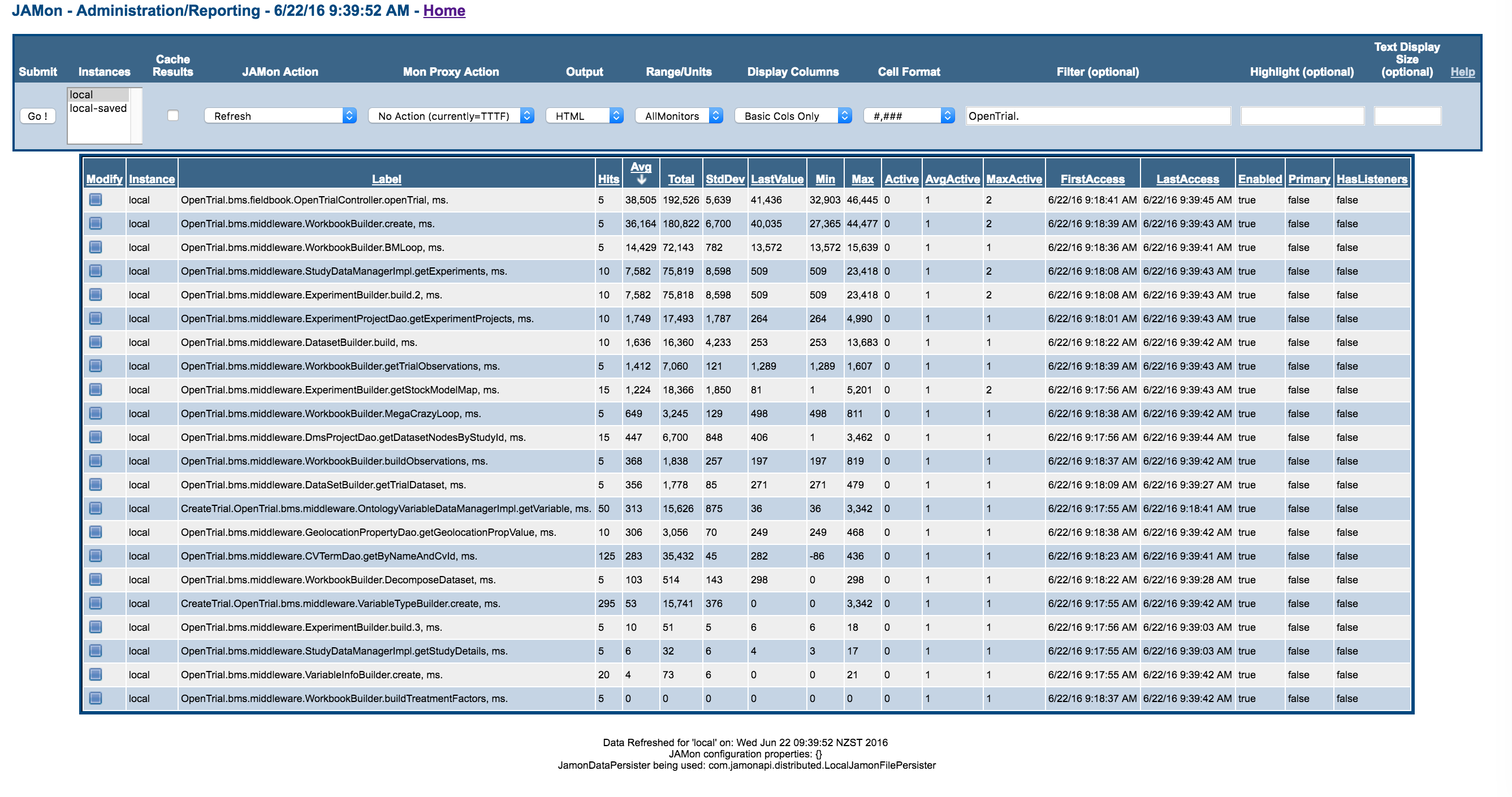
Profiling observations
For trial with 200 locations (each with one location attribute and one condition), 25 traits, 3 reps, 50 germplasm entries. One environment containing complete measurements = 150 x 25 = 3750 phenotypes.
Backend
- Average time spent on the backend : 38,505ms (WorkbookBuilder). Breakdown of time spent:
- 14,429ms (~37%) spent in processing to handle "special" variables that represent Breeding Methods.
- Large remaining portion spent on retrieving observation units (nd_experiment) germplasm (stock) and related join tables and generic properties tables. Query volume is directly proportional to number of observation units with multiple extra rows per unit as meta data in props table.
UI
- Average time to render the page : 1.9 minutes. Most of this time is spent in rendering the measurement data table UI.
- After configuring deferRender property of data table, the time taken reduces down to 15 seconds
Recommendations for discussion
- Fix the modelling of Breeding Methods as ontology variables (after fixing this average backend time drops to ~25 Sec).
- Do not load information about all observation units, traits and phenotypes on screen. Currently this is being done to display the entire measurements table on screen which is a major contributor to performance issues with large trials.
- Limit the number of observation units displayed.
- Consider server side pagination. Sorting by traits with server side pagination is going to be extremely complicated with current schema structure.
- Only show certain number of observations and associated phenotypes on screen. Beyond a fixed limit, use excel export to see all data. Depends on how the breeders actually want to interact with and use the whole trial data.
- Consider a completely different way of visualising the measurement table data.
Import Export Measurements
The Import Measurements flow shows no significant time spent importing measurements. The bottlenecks occur inside the Save Nursery/Trial section of the product, which is covered in the Create/Open Trial section of this page.
The BMS is littered with transformations, suggesting that we have
- mismatch in the data structures we transpose throughout the application
- semi-generic structures that need filtering and refining The Breeding Method codes are found in the Methods tables, but the keys that identify these codes are varied and stored in the Ontology. Switching between ids, names and codes for Breeding Method at the plot level is costing us dearly in performance pitfalls in the code.
Open Trial Query Improvements
Open trial performance can be further optimised by writing a complex query. Below is a query that retrieves all measurement related information for a very large trial in a couple of seconds.
SET SESSION group_concat_max_len = 1000000;
set @sql = NULL;
SELECT
GROUP_CONCAT(DISTINCT
CONCAT(
'Max(IF(cvterm_variable.name = ''',
cvt.name,
''', ph.value, NULL)) AS ',
cvt.name
)
) INTO @sql
FROM
projectprop pp
INNER JOIN
cvterm cvt ON cvt.name = pp.value
WHERE
type_id = 1808
AND project_id = (SELECT
p.project_id
FROM
project_relationship pr
INNER JOIN
project p ON p.project_id = pr.subject_project_id
WHERE
pr.object_project_id = 2007
AND name LIKE '%PLOTDATA');
SET @sql = CONCAT('SELECT
nde.nd_experiment_id,
gl.description AS TRIAL_INSTANCE,
(SELECT
iispcvt.definition
FROM
stockprop isp
INNER JOIN
cvterm ispcvt ON ispcvt.cvterm_id = isp.type_id
INNER JOIN
cvterm iispcvt ON iispcvt.cvterm_id = isp.value
WHERE
isp.stock_id = s.stock_id
AND ispcvt.name = ''ENTRY_TYPE'') ENTRY_TYPE,
s.dbxref_id AS GID,
s.name DESIGNATION,
s.uniquename ENTRY_NO,
(SELECT
isp.value
FROM
stockprop isp
INNER JOIN
cvterm ispcvt1 ON ispcvt1.cvterm_id = isp.type_id
WHERE
isp.stock_id = s.stock_id
AND ispcvt1.name = ''SEED_SOURCE'') SEED_SOURCE,
(SELECT
ndep.value
FROM
nd_experimentprop ndep
INNER JOIN
cvterm ispcvt ON ispcvt.cvterm_id = ndep.type_id
WHERE
ndep.nd_experiment_id = ep.nd_experiment_id
AND ispcvt.name = ''REP_NO'') REP_NO,
(SELECT
ndep.value
FROM
nd_experimentprop ndep
INNER JOIN
cvterm ispcvt ON ispcvt.cvterm_id = ndep.type_id
WHERE
ndep.nd_experiment_id = ep.nd_experiment_id
AND ispcvt.name = ''PLOT_NO'') PLOT_NO,',@sql,
' FROM
Project p
INNER JOIN
project_relationship pr ON p.project_id = pr.subject_project_id
INNER JOIN
nd_experiment_project ep ON pr.subject_project_id = ep.project_id
INNER JOIN
nd_experiment nde ON nde.nd_experiment_id = ep.nd_experiment_id
INNER JOIN
nd_geolocation gl ON nde.nd_geolocation_id = gl.nd_geolocation_id
INNER JOIN
nd_experiment_stock es ON ep.nd_experiment_id = es.nd_experiment_id
INNER JOIN
Stock s ON s.stock_id = es.stock_id
LEFT JOIN
nd_experiment_phenotype neph ON neph.nd_experiment_id = nde.nd_experiment_id
LEFT JOIN
phenotype ph ON neph.phenotype_id = ph.phenotype_id
LEFT JOIN
cvterm cvterm_variable ON cvterm_variable.cvterm_id = ph.observable_id
WHERE
p.project_id = (Select p.project_id from project_relationship pr
INNER JOIN project p on p.project_id = pr.subject_project_id
where (pr.object_project_id = 2007 and name LIKE ''%PLOTDATA''))
GROUP BY nde.nd_experiment_id, TRIAL_INSTANCE, ENTRY_TYPE, SEED_SOURCE, REP_NO, PLOT_NO
');
PREPARE stmt FROM @sql;
EXECUTE stmt;
DEALLOCATE PREPARE stmt;
# SHOW VARIABLES;
#
Obviously, this query can be further optimised but provides a flavour of what's possible by optimising our query.
Advance
Advancing is hampered only by the unique name checking for Germplasm as is required in most systems. These candidate names should first be 'allocated' from a Name Registry that can discern for a batch of records up front as to whether a range of names are unique.
Germplasm Search
Key Stats
- Total Germplasm Search took 2513 ms
- Count Query took - 50 ms
- Search for germplasm took - 38 ms
- Pedigree Generation Took - 1853 ms i.e. 73% percent of the total processing time.
Recommendation
- Optimise Pedigree String retrieval be storing the pedigree string
- Lazy generation of pedigree string.
- Batch processor to update pedigree string when Pedigree string is invalidated.
- Possible database optimisations by moving the selection history out of the germplasm table.
- Alternative approaches to storing ancestry tree
Interesting finding
Retrieval of the pedigree tree could have been faster if MySQL supported recursive queries. Unfortunately, MySql does not. We can work around this with using a stored procedure.
# Usage: the standard syntax:
# WITH RECURSIVE recursive_table AS
# (initial_SELECT
# UNION ALL
# recursive_SELECT)
# final_SELECT;
# should be translated by you to
# CALL WITH_EMULATOR(recursive_table, initial_SELECT, recursive_SELECT,
# final_SELECT, 0, "").
# ALGORITHM:
# 1) we have an initial table T0 (actual name is an argument
# "recursive_table"), we fill it with result of initial_SELECT.
# 2) We have a union table U, initially empty.
# 3) Loop:
# add rows of T0 to U,
# run recursive_SELECT based on T0 and put result into table T1,
# if T1 is empty
# then leave loop,
# else swap T0 and T1 (renaming) and empty T1
# 4) Drop T0, T1
# 5) Rename U to T0
# 6) run final select, send relult to client
# This is for *one* recursive table.
# It would be possible to write a SP creating multiple recursive tables.
delimiter |
CREATE PROCEDURE WITH_EMULATOR(
recursive_table varchar(100), # name of recursive table
initial_SELECT text, # seed a.k.a. anchor
recursive_SELECT text, # recursive member
final_SELECT text, # final SELECT on UNION result
max_recursion int unsigned, # safety against infinite loop, use 0 for default
create_table_options text # you can add CREATE-TABLE-time options
# to your recursive_table, to speed up initial/recursive/final SELECTs; example:
# "(KEY(some_column)) ENGINE=MEMORY"
)
BEGIN
declare new_rows int unsigned;
declare show_progress int default 0; # set to 1 to trace/debug execution
declare recursive_table_next varchar(120);
declare recursive_table_union varchar(120);
declare recursive_table_tmp varchar(120);
set recursive_table_next = concat(recursive_table, "_next");
set recursive_table_union = concat(recursive_table, "_union");
set recursive_table_tmp = concat(recursive_table, "_tmp");
# Cleanup any previous failed runs
SET @str =
CONCAT("DROP TEMPORARY TABLE IF EXISTS ", recursive_table, ",",
recursive_table_next, ",", recursive_table_union,
",", recursive_table_tmp);
PREPARE stmt FROM @str;
EXECUTE stmt;
# If you need to reference recursive_table more than
# once in recursive_SELECT, remove the TEMPORARY word.
SET @str = # create and fill T0
CONCAT("CREATE TEMPORARY TABLE ", recursive_table, " ",
create_table_options, " AS ", initial_SELECT);
PREPARE stmt FROM @str;
EXECUTE stmt;
SET @str = # create U
CONCAT("CREATE TEMPORARY TABLE ", recursive_table_union, " LIKE ", recursive_table);
PREPARE stmt FROM @str;
EXECUTE stmt;
SET @str = # create T1
CONCAT("CREATE TEMPORARY TABLE ", recursive_table_next, " LIKE ", recursive_table);
PREPARE stmt FROM @str;
EXECUTE stmt;
if max_recursion = 0 then
set max_recursion = 100; # a default to protect the innocent
end if;
recursion: repeat
# add T0 to U (this is always UNION ALL)
SET @str =
CONCAT("INSERT INTO ", recursive_table_union, " SELECT * FROM ", recursive_table);
PREPARE stmt FROM @str;
EXECUTE stmt;
# we are done if max depth reached
set max_recursion = max_recursion - 1;
if not max_recursion then
if show_progress then
select concat("max recursion exceeded");
end if;
leave recursion;
end if;
# fill T1 by applying the recursive SELECT on T0
SET @str =
CONCAT("INSERT INTO ", recursive_table_next, " ", recursive_SELECT);
PREPARE stmt FROM @str;
EXECUTE stmt;
# we are done if no rows in T1
select row_count() into new_rows;
if show_progress then
select concat(new_rows, " new rows found");
end if;
if not new_rows then
leave recursion;
end if;
# Prepare next iteration:
# T1 becomes T0, to be the source of next run of recursive_SELECT,
# T0 is recycled to be T1.
SET @str =
CONCAT("ALTER TABLE ", recursive_table, " RENAME ", recursive_table_tmp);
PREPARE stmt FROM @str;
EXECUTE stmt;
# we use ALTER TABLE RENAME because RENAME TABLE does not support temp tables
SET @str =
CONCAT("ALTER TABLE ", recursive_table_next, " RENAME ", recursive_table);
PREPARE stmt FROM @str;
EXECUTE stmt;
SET @str =
CONCAT("ALTER TABLE ", recursive_table_tmp, " RENAME ", recursive_table_next);
PREPARE stmt FROM @str;
EXECUTE stmt;
# empty T1
SET @str =
CONCAT("TRUNCATE TABLE ", recursive_table_next);
PREPARE stmt FROM @str;
EXECUTE stmt;
until 0 end repeat;
# eliminate T0 and T1
SET @str =
CONCAT("DROP TEMPORARY TABLE ", recursive_table_next, ", ", recursive_table);
PREPARE stmt FROM @str;
EXECUTE stmt;
# Final (output) SELECT uses recursive_table name
SET @str =
CONCAT("ALTER TABLE ", recursive_table_union, " RENAME ", recursive_table);
PREPARE stmt FROM @str;
EXECUTE stmt;
# Run final SELECT on UNION
SET @str = final_SELECT;
PREPARE stmt FROM @str;
EXECUTE stmt;
# No temporary tables may survive:
SET @str =
CONCAT("DROP TEMPORARY TABLE ", recursive_table);
PREPARE stmt FROM @str;
EXECUTE stmt;
# We are done :-)
END|
delimiter ;
Example usage with the germplasm table
CALL WITH_EMULATOR (
"germplsm_recursive",
"
SELECT *
FROM germplsm g
where g.gid = 500012",
"
SELECT kid.* from germplsm kid
INNER join germplsm p ON p.gpid1 = kid.gid or p.gpid2 = kid.gid
where p.gid in (Select gid from germplsm_recursive)
",
"
SELECT *
FROM germplsm_recursive;
",
10000000,
"ENGINE=MEMORY"
) ;
This workaround would have been great but this does not work in a transactional system as the creation of tempory tables effectively kills the transactions.