Xmipp release notes - I2PC/scipion GitHub Wiki
We are very pleased to announce the release of a new version of Xmipp. It’s been over a year since the previous and we have been working on new EM methods and improving Scipion as well. Some of the new developments are listed below.
Adds noise to particles or volumes of different types: Uniform, Student or Gaussian.
Applies transformation matrix of an aligned volume on a set of particles to modify their angular assignment.
Creates a gallery of projections from a volume. This gallery of projections may help to understand the images observed in the microscope.
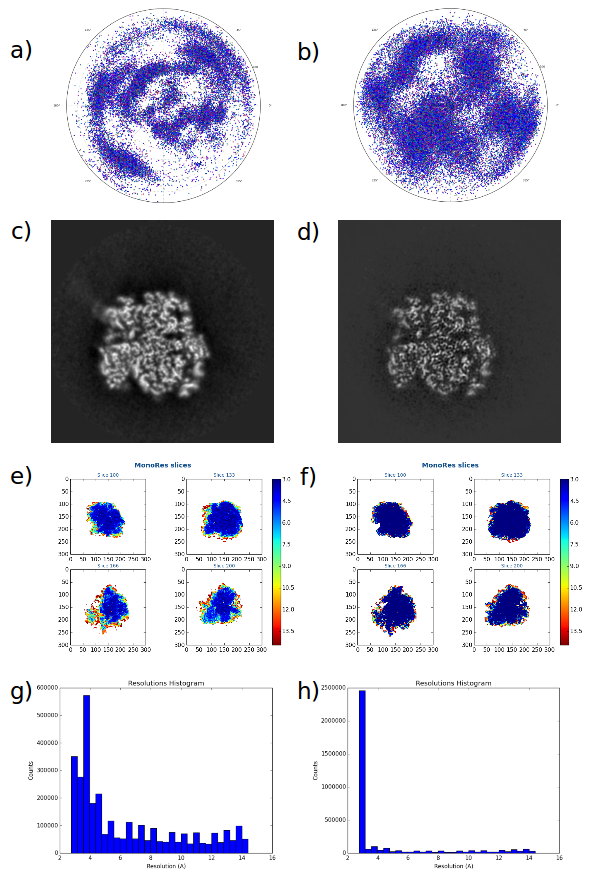
A new 3D refinement protocol for solving the problem of reconstructing the 3D structure of a macromolecule from an homogeneous population of images. It is based on a subproblem decomposition:
-
Angular assignment in which an experimental image can occupy multiple orientations;
-
3D Reconstruction with weights;
-
Volume restoration. The algorithm is normally used in the so-called "gold-standard" paradigm. However, this is not a requirement of the algorithm.
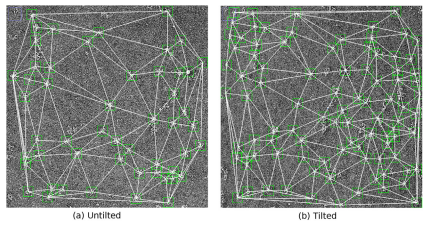
Random Conical Tilt requires establishing the correspondence between untilted and tilted particles. This protocol allows automatic picking on the untilted, automatic picking on the tilted and the protocol will automatically find the correspondences. See the paper here.
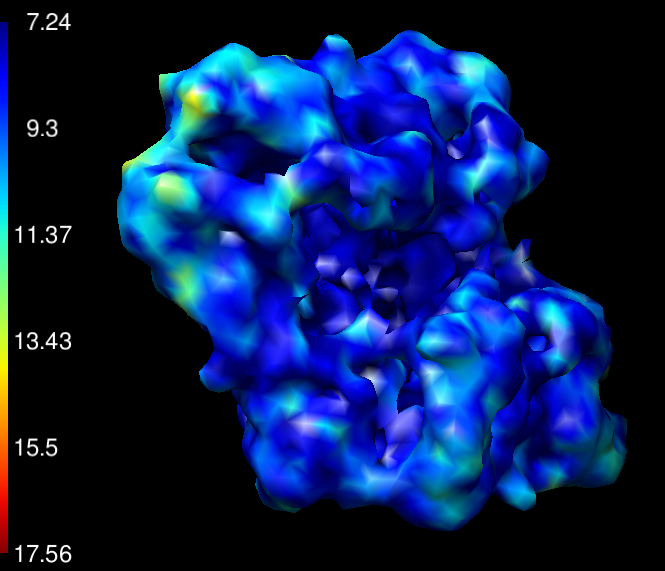
local monoRes: local resolution estimation
Estimates the local resolutions to each voxel of a given map or half maps.
Estimates the gain image of a DDD camera, directly analyzing the movies. This gain is needed to have the set of recorded frames into a coherent gray level range, homogeneous over the whole image. The algorithm does not need any other input than the DED movie itself, being capable of providing an estimate of the camera gain image, helping to identify dead pixels and cases of incorrectly calibrated cameras. We propose the algorithm to be used either to validate the experimentally acquired gain image (for instance, to follow its possible change over time) or to verify that there is no residual gain image after experimentally correcting for the camera gain.

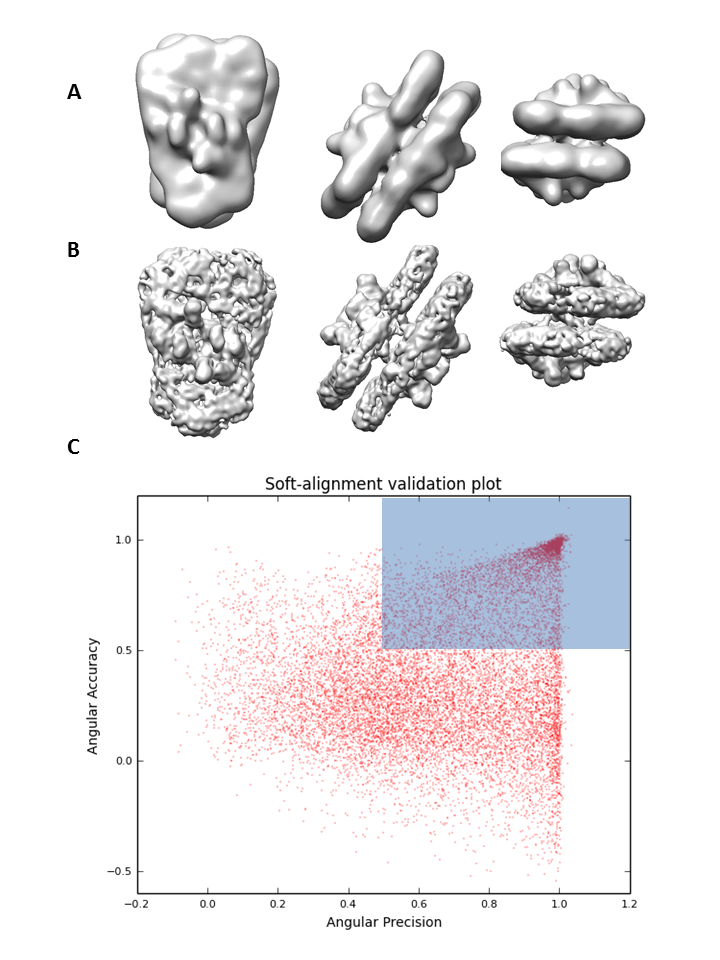
Performs soft alignment validation of a set of particles confronting them against a given 3DEM map, estimating the alignment precision and accuracy for each particle. This approach is able to quantify the alignment precision and accuracy of the 3D alignment process, which is then being used to help the reconstruction process in a number of ways, such as:
-
Providing quality indicators of the macromolecular map for soft validation
-
Assessing the degree of homogeneity of the sample
-
Selecting subsets of representative images (pruning).
Given two states of the same molecule, calculate the local forces that deform one state into the other. The analysis of these forces reveal local rotations and local stretches/compressions. See the paper here.
Construct image groups based on the angular assignment. All images assigned within a solid angle are assigned to a class. Classes are not exclusive and an image may be assigned to multiple classes.
A quantitative analysis of dissimilarities (distances) among the EM maps that placing the entire set of density maps into a common space of comparison.The approach is based on statistical analysis of distance among elastically aligned EM maps, and results in visualizing those maps as points in a lower dimensional distance space. See the paper here.
Checks how the resolution changes with the number of projections used for 3D reconstruction. This method has been proposed by B. Heymann "Validation of 3D EM Reconstructions", 2015.