Variant DB Architecture - GenomicsDB/GenomicsSampleAPIs GitHub Wiki
High Level System Architecture
General Info
VariantDB (aka Variant Store) is a GA4GH interface which leverages a MetaDB and GenomicsDB (TileDB optimized for variant storage and access). For more information on GenomicsDB, how it relates to TileDB, and common terminology see here. More information on MetaDB is at the bottom of this page.
Information Flow of a VariantDB Query
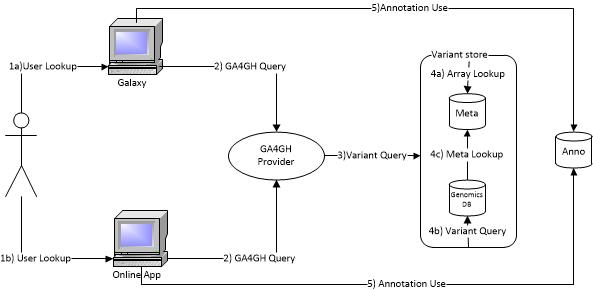
###1. User query about variant
-
A user has a question to ask about variants. They generate the query and ask through GA4GH REST API, or an Online query system that uses the GA4GH API.
-
Example:
Select all variants that fall on chromosome 1 for hg19 genome.
###2. Send GA4GH query
-
Whether generated by the user, or by their tool, a GA4GH query is sent to our GA4GH provider (Flask / Python interface)
-
Example (con't):
curl -H "Content-Type: application/json" -X POST -d {'start': 1, 'end': 249250621, 'referenceName': 'chr1'} http://localhost:8008/variants/search
###3. Variant Query
- The GA4GH interface of the Variant Store consolidates the information needed to construct an appropriate GA4GH request for the given Variant Store instance (MetaDB, GenomicsDB, etc.).
###4. Query Handling
####4a. Lookup Workspace and Array information from MetaDB to be used in GenomicsDB query
- The GA4GH backend does a search against MetaDB to retrieve information that is required to perform a query to a TileDB array. This information is consolidated along with the columns (genomic region) and rows (callsets) in question and used to perform set in 4b.
####4b. Query GenomicsDB for Variant Information
- The translated query from 4a is sent to the TileDB backend. The data is collected, filtered, and constructed while building the whole GA4GH response that is finalized in step 4c.
####4c. Meta data lookup
-
The variants that are returned from TileDB need to be mapped back to their respective GA4GH identities, not stored in GenomicsDB for efficiency reasons.
-
Example (con't):
The posted json is translated into a region query (4a), this is sent to GenomicsDB which returns a set of variants (4b). This set of variants is sent back to the user as a GA4GH response by pulling additional data about CallSets and VariantSets from the MetaDB (4c).
###5. Apply external annotation information
- The online app, galaxy tool, or user calls the Annotation system on the resultant data to obtain the annotated result (if required).
MetaDB
MetaDB Models
MetaDB is suited to work with postgres. The models below are an integration between GA4GH schemas and models that support handling general metadata for GenomicsDB. The data model for MetaDB is below:

MetaDB API
The MetaDB API includes:
- DBImport: class to populate a MetaDB instance upon importing a set of variants into a TileDB array.
- Query: class to query a MetaDB instance to interpret a GA4GH request, communicate with TileDB, and construct a GA4GH response.