Home - GenomicSEM/GenomicSEM GitHub Wiki
Welcome to the genomicSEM wiki!
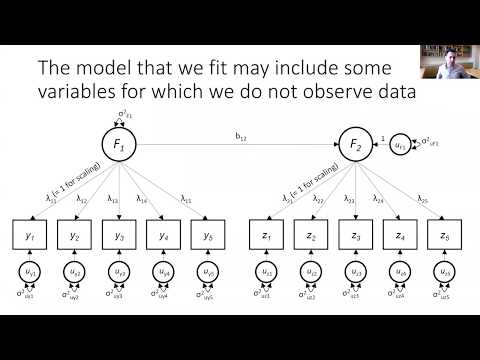
GenomicSEM is software to estimate a structural equation model on traits, for which you only have access to GWAS summary statistics.
There are two primary types of models in GenomicSEM: one that includes the effect of individual SNPs and one that does not include SNP effects. Please see the tutorials for Models with SNP Effects and Models without SNP effects for details on how to run each. Answers to common questions can be found on the FAQs page (which we will launch as soon as we frequently get actual questions). If you are having issues and not finding the answers anywhere on the wiki or FAQs page, we encourage you to post your question on the google group.
Code Updates: The two most recent code updates are to userGWAS. These reflect default behavior to fix the measurement model across SNPs and to automatically calculate Q_SNP for any factor that is predicted by a SNP (without needing to estimate a follow-up independent pathways model).
On September 23rd, 2021 we updated the munge function and wiki to use the sum of effective sample sizes for performing the liability h^2 conversion for binary traits. This is based on simulation findings from our group that indicate this is the more appropriate way to perform the liability-scale conversion as using total sample size and observed sample prevalence (the prior field standard) can cause substantial downward bias in estimates. (https://www.medrxiv.org/content/10.1101/2021.09.22.21263909v1).
The prior update is dated April 30th, 2021 and includes the documentation and functionality to run Transcriptome-wide Structural Equation Models. The prior code update, dated April 12th, 2021, includes a statistically equivalent, but far more efficient, way of calculating model chi-square. In the context of a userGWAS model where model chi-square was requested this may decrease run-times by ~50%. We recommend reinstalling to the most recent update of GenomicSEM. However, please note that changes in Genomic SEM defaults are likely to produce slight changes in results relative to previous versions. For further details, see version history
Introduction of Stratified Genomic SEM: Stratified Genomic SEM can now be used to examine enrichment for any parameter for a model estimating in Genomic SEM.
Introduction of Transcriptome-wide SEM T-SEM can be used to examine the effect of imputed gene expression from FUSION on a common factor indexing genetic sharing across correlated traits.
Feature update: GenomicSEM can now run HDL a novel method for estimating heritability and genetic correlation that can in some cases outperform LDSC. See out tutorial HERE
Contents of the wiki:
Learn how to install GenomicSEM
Consider some of the nuances of summary data, and know where to find summary data.
Fit SEM models to GWAS summary data without a SNP
Run a GWAS where the SNP is included in the structural equation model.
Run a GWAS with the SNP effect(s) included in a user specified SEM
Estimate functional enrichment for any parameter in a Genomic SEM model (e.g., factor variances).
Run multivariate TWAS using T-SEM
Get guidance on advanced models on this page with tutorials which showcases and explains specific structural equation models you can fit using GenomicSEM
PGC worldwide lab meeting on genomicSEM
Click below for a video which provides an introduction to the method/paper: