EMxtalinfo - EMsoft-org/EMsoft GitHub Wiki
Program: EMxtalinfo
This utility program combines a few other programs into a single program. The user is asked to select a crystal structure, microscope accelerating voltage, and scattering factor set. Zone axis diffraction patterns require entry of the camera length (in mm), a PostScript output file name, a maximum index value for stereographic projections, logarithmic or exponential intensity scaling for the kinematical diffraction patterns, and selected zone axes. This program is typically useful for TEM observations, to assist the user while working at the microscope.
A typical usage example would be the following
...
Crystallographic Information
Structure Factors
Total number of families = 35
Total number of family members = 694
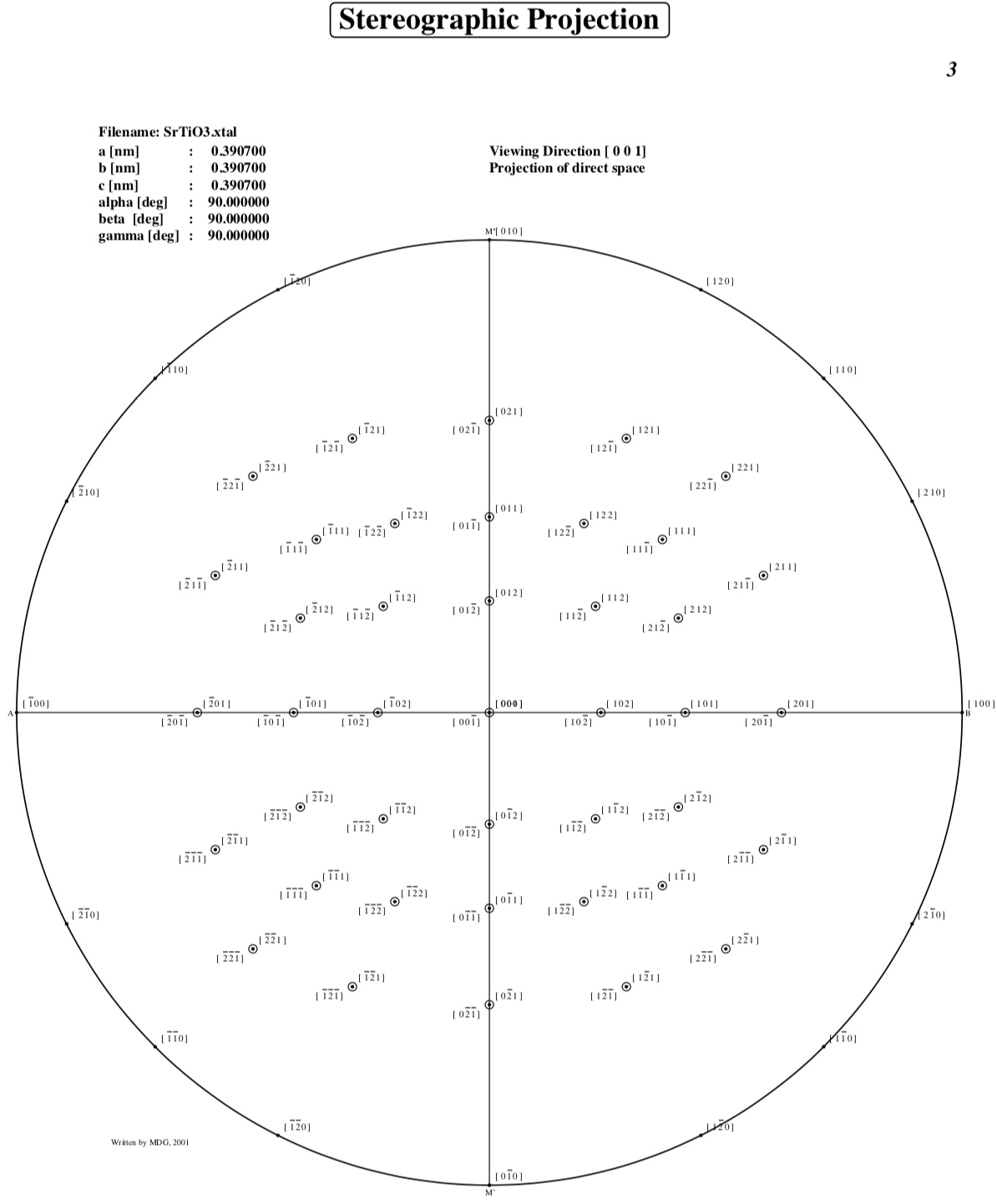
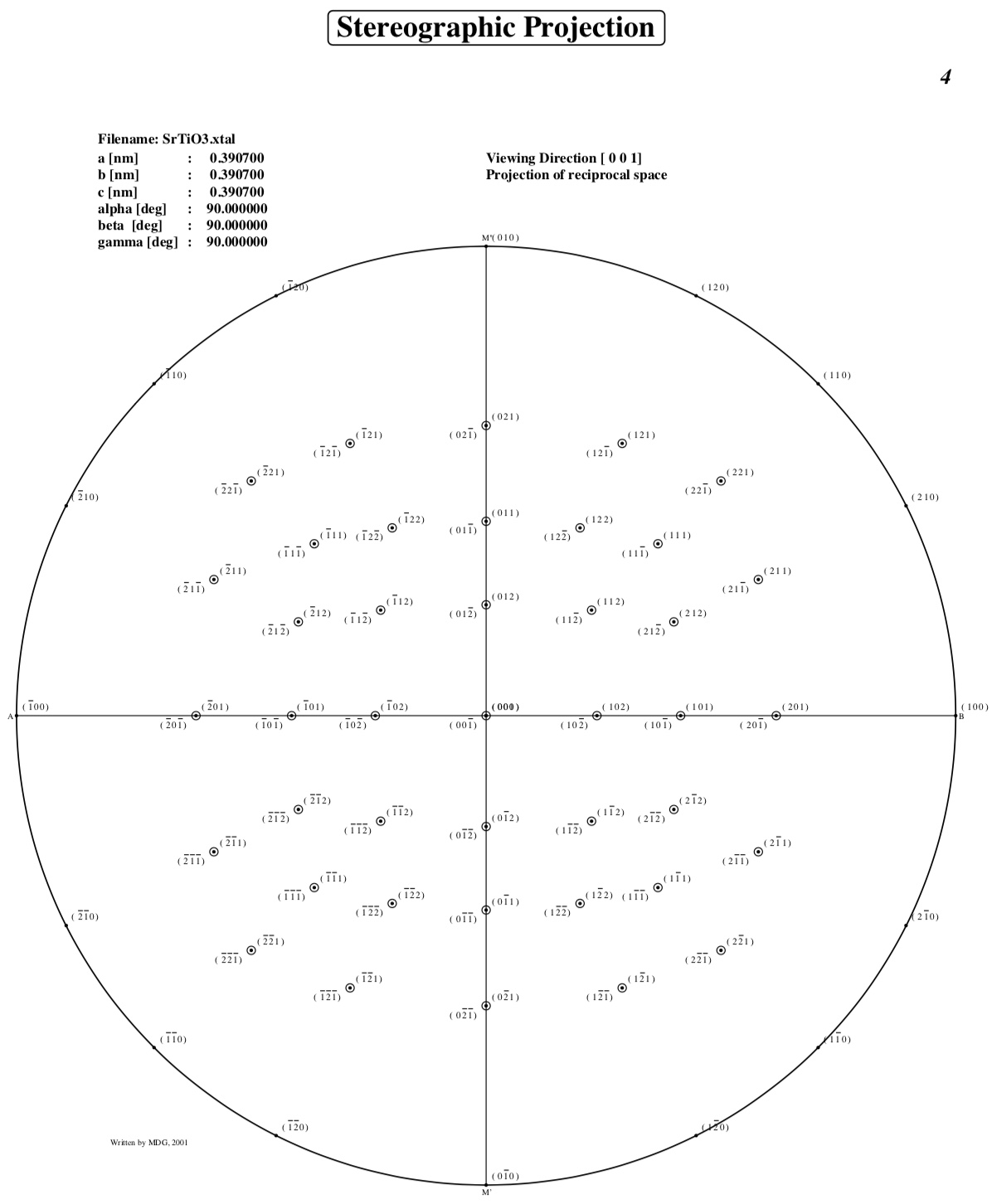
Stereographic Projections
Enter the maximum index for h,k and l, or for
u,v, and w. For a hexagonal system, please use
4-index notation [uv.w] or (hk.l) to determine
the largest index.
Enter maximum indices : 2,2,2
Diffraction Patterns
wavelength [nm] = 2.50777137E-03
L [mm] = 1200.00000
camera length lambda*L [mm nm] = 3.00932574
logarithmic[0] or exponential[1] intensity scale 1
->Total number of direction families = 40
List of available zone axis patterns
1 [ 1 0 0]; 2 [ 1 1 0]; 3 [ 1 1 1]; 4 [ 2 1 0];
5 [ 2 1 1]; 6 [ 2 2 1]; 7 [ 3 1 0]; 8 [ 3 1 1];
9 [ 3 2 0]; 10 [ 3 2 1]; 11 [ 3 2 2]; 12 [ 4 1 0];
13 [ 4 1 1]; 14 [ 3 3 1]; 15 [ 4 2 1]; 16 [ 3 3 2];
17 [ 4 3 0]; 18 [ 5 1 0]; 19 [ 4 3 1]; 20 [ 5 1 1];
21 [ 4 3 2]; 22 [ 5 2 0]; 23 [ 5 2 1]; 24 [ 5 2 2];
25 [ 4 4 1]; 26 [ 4 3 3]; 27 [ 5 3 0]; 28 [ 5 3 1];
29 [ 5 3 2]; 30 [ 5 4 0]; 31 [ 4 4 3]; 32 [ 5 4 1];
33 [ 5 3 3]; 34 [ 5 4 2]; 35 [ 5 4 3]; 36 [ 5 5 1];
37 [ 5 5 2]; 38 [ 5 4 4]; 39 [ 5 5 3]; 40 [ 5 5 4];
Enter selection (e.g. 4,10-20, ... )
[Include 0 to also draw a powder pattern]
-> 1-12
No indices (0), labels (1), extinctions (2), labels + extinctions (3): 1
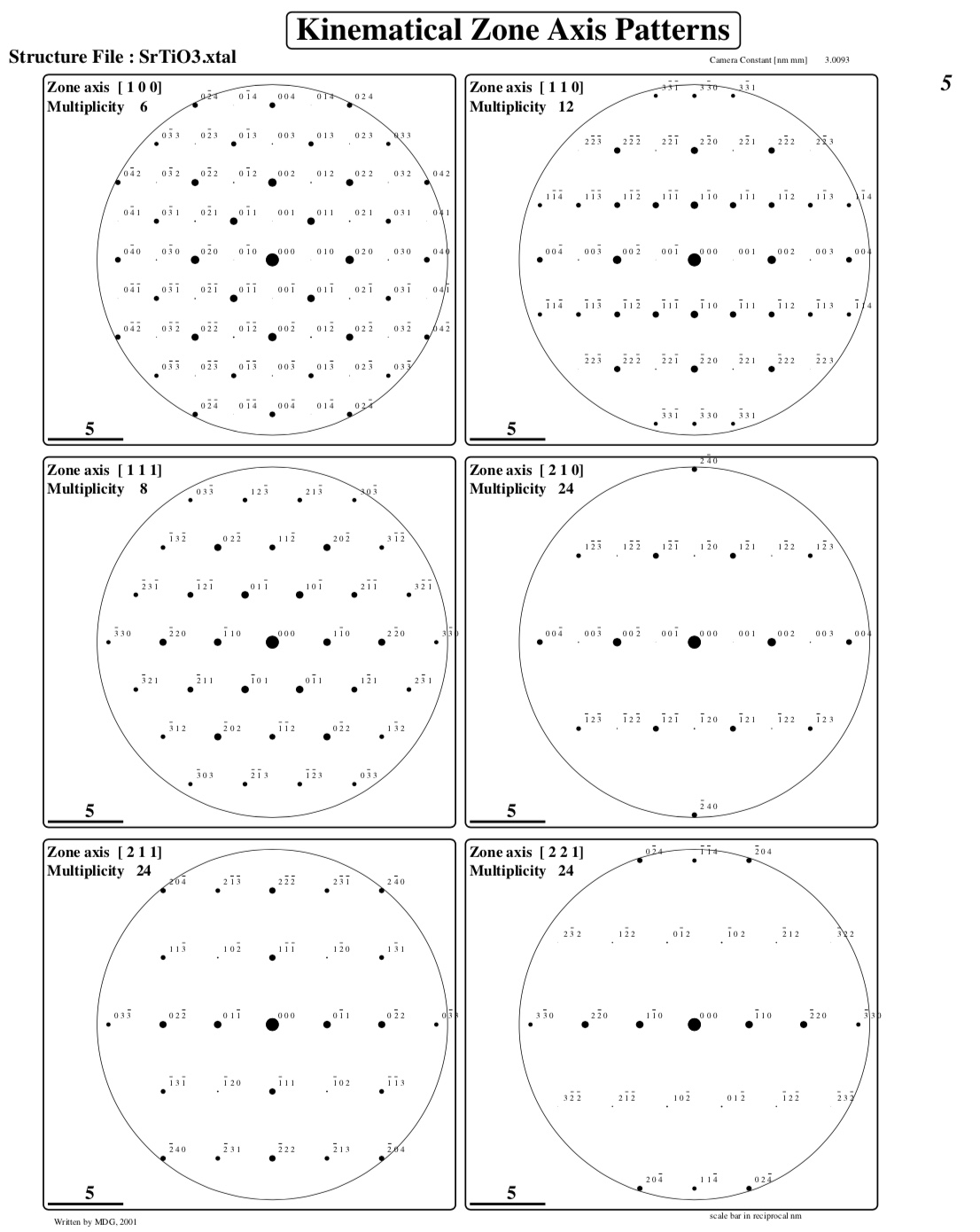
Creating ZAP[ 1 0 0] : Number of reflections : 69
Creating ZAP[ 1 1 0] : Number of reflections : 47
Creating ZAP[ 1 1 1] : Number of reflections : 37
Creating ZAP[ 2 1 0] : Number of reflections : 25
Creating ZAP[ 2 1 1] : Number of reflections : 27
Creating ZAP[ 2 2 1] : Number of reflections : 25
Creating ZAP[ 3 1 0] : Number of reflections : 23
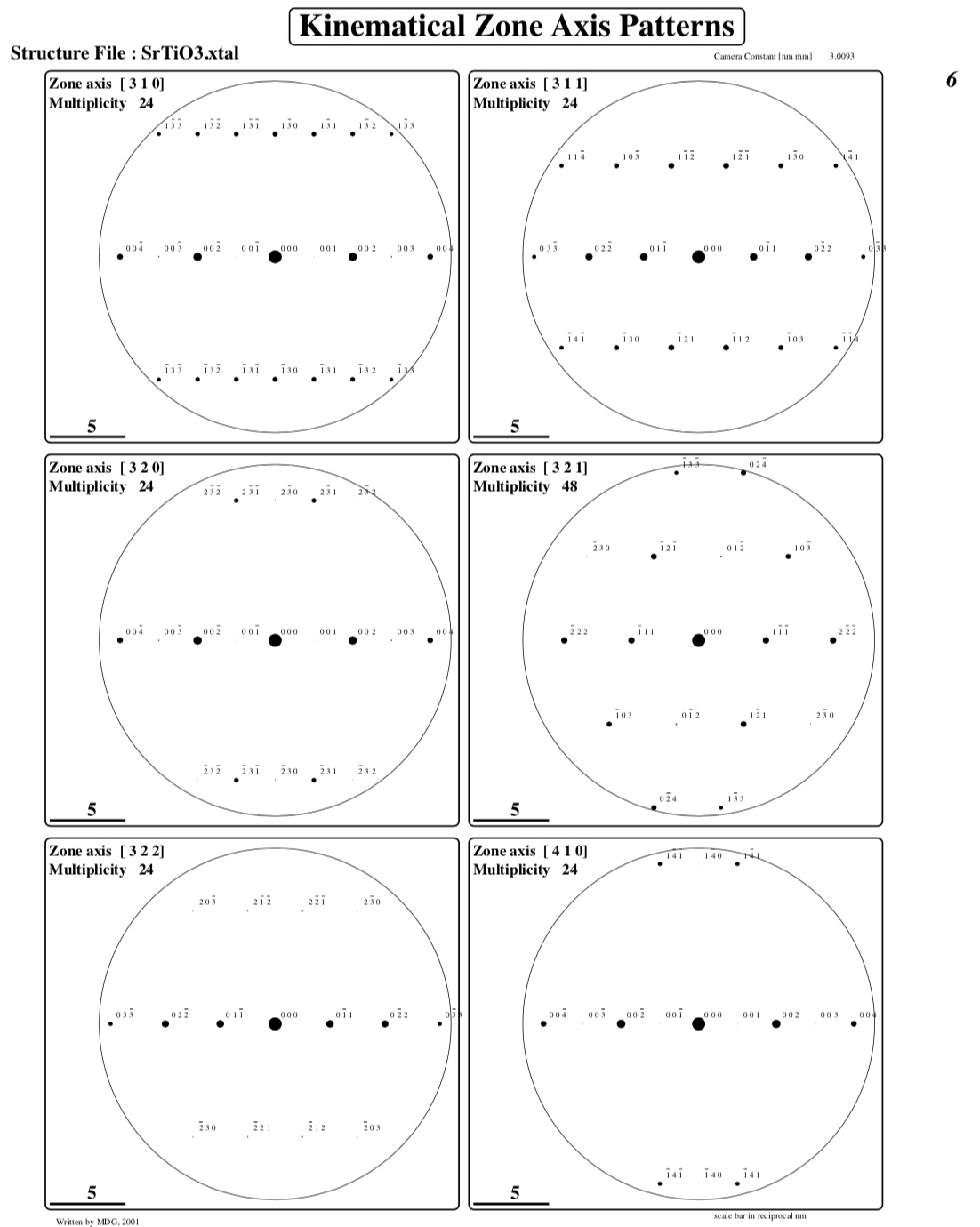
Creating ZAP[ 3 1 1] : Number of reflections : 19
Creating ZAP[ 3 2 0] : Number of reflections : 19
Creating ZAP[ 3 2 1] : Number of reflections : 17
Creating ZAP[ 3 2 2] : Number of reflections : 15
Creating ZAP[ 4 1 0] : Number of reflections : 15
The output is a PostScript file with several pages (6, in this case):
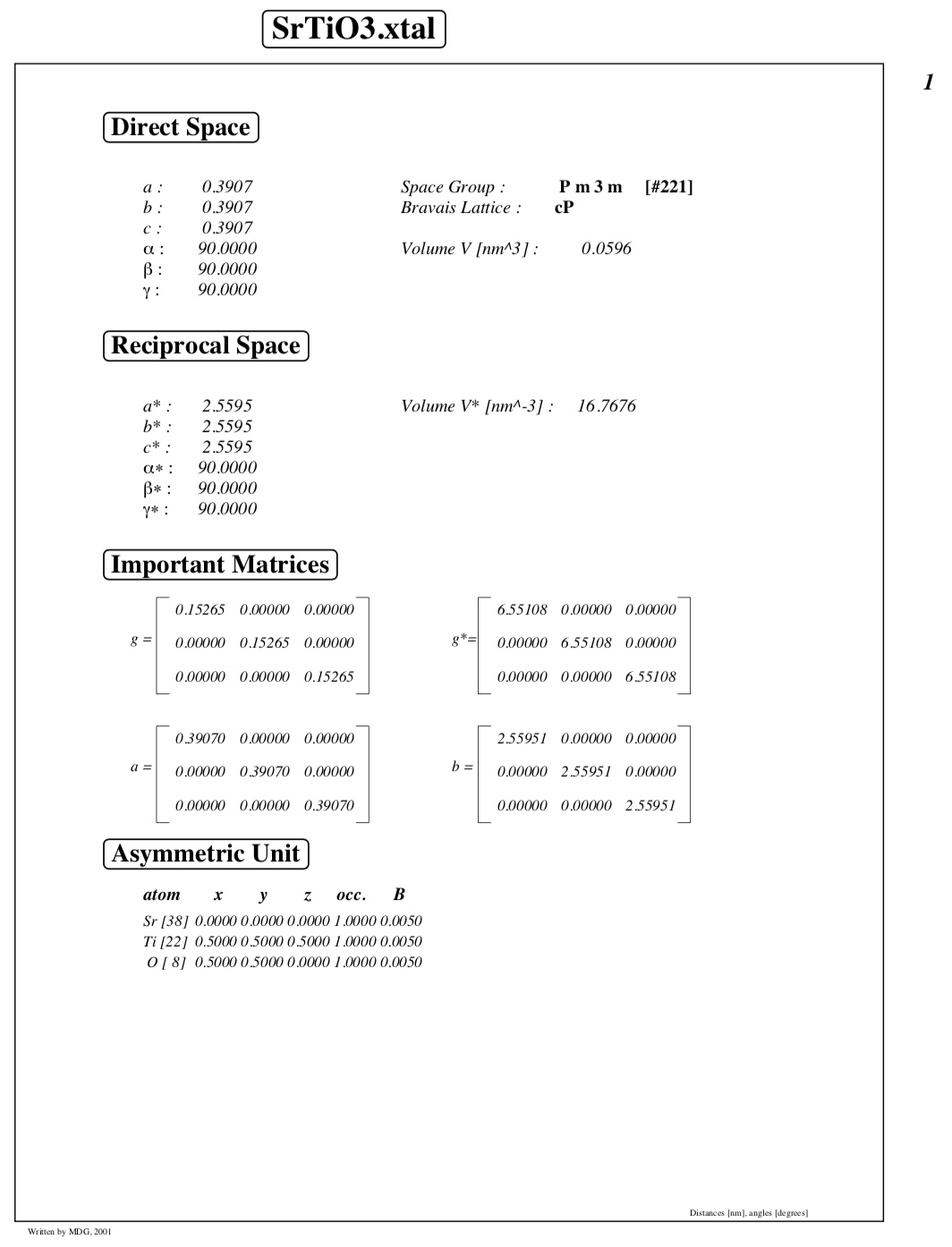
Crystal structure information, including atom coordinates and metric tensors/structure matrices:
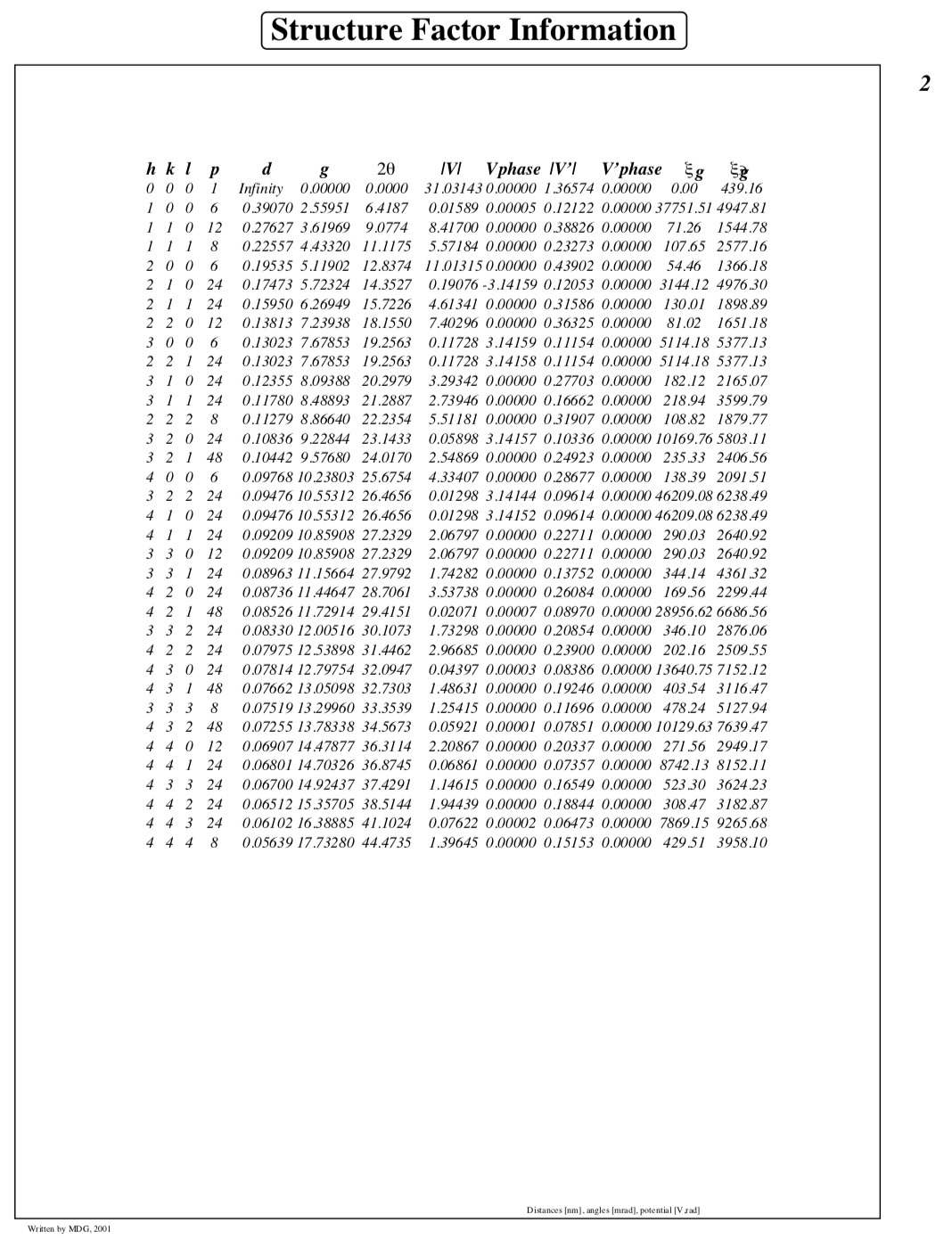
Fourier coefficients of the lattice potential along with extinction distances and absorption lengths:
Direct space stereographic projection:
Reciprocal space stereographic projection:
One or more pages of kinematical diffraction patterns: