Home - DianaCarolinaVergara/SNPs_pipeline GitHub Wiki
Welcome to the SNPs_pipeline wiki!
Phylogeny Construction with SNPs
Pipeline
We received a vcf file where SNPsaurus converted genomic DNA into nextRAD genotyping-by-sequencing libraries (SNPsaurus, LLC).
The nextRAD libraries were sequenced on a HiSeq 4000 on two lanes of 150bp reads.
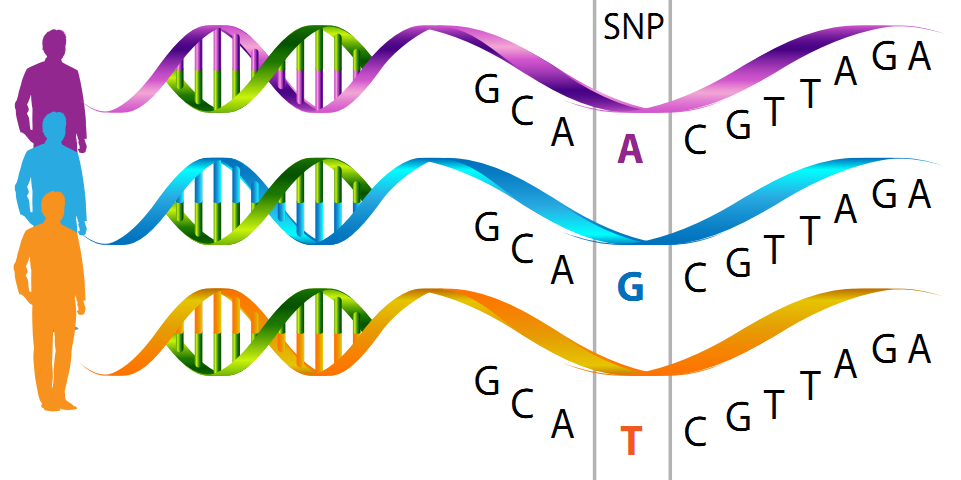
With this pipeline you would be able to:
Modified from http://grunwaldlab.github.io/Population_Genetics_in_R/qc.html
- Using RStudio 3.4.4 and packages:
genepop, parallel, poppr, dartR, devtools, phytools, seqinr, phylotools, adegenet, pegas, hierfstat
-
Remove
indels(insertions-deletions) -
SNPs upper and lower 20% of depth distribution
-
Delete:
3.1 Samples (missingness >70%)
3.2 SNPs (>90%) with a high degree of missingness information
-
Rewrite
vcf file
-
Using RStudio or VCFTOOLS 0.1.17
- Run Minor Allele Frequency (MAF)
-
Convert vcf file to
PHYLIP format
Suitable for RAxML
Or you can use another tool from CIPRESS Phylogenetic Collection (BEAST2, MRBAYES, RAXML) CyVerse

References
-
http://grunwaldlab.github.io/Population_Genetics_in_R/qc.html
-
RStudio: A Platform‐Independent IDE for R and Sweave - Racine - 2012 - Journal of Applied Econometrics - Wiley Online Library. https://onlinelibrary.wiley.com/doi/abs/10.1002/jae.1278.
-
variant call format and VCFtools | Bioinformatics | Oxford Academic. https://academic.oup.com/bioinformatics/article/27/15/2156/402296.
-
Lischer, H. E. L. & Excoffier, L. PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28, 298–299 (2012).
-
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
-
Miller, M. A., Pfeiffer, W. & Schwartz, T. The CIPRES Science Gateway: Enabling High-impact Science for Phylogenetics Researchers with Limited Resources. in Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the eXtreme to the Campus and Beyond 39:1–39:8 (ACM, 2012). doi:10.1145/2335755.2335836.
-
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).

