4. Quality Filter - DianaCarolinaVergara/16S-rRNA-Analysis GitHub Wiki
DADA2
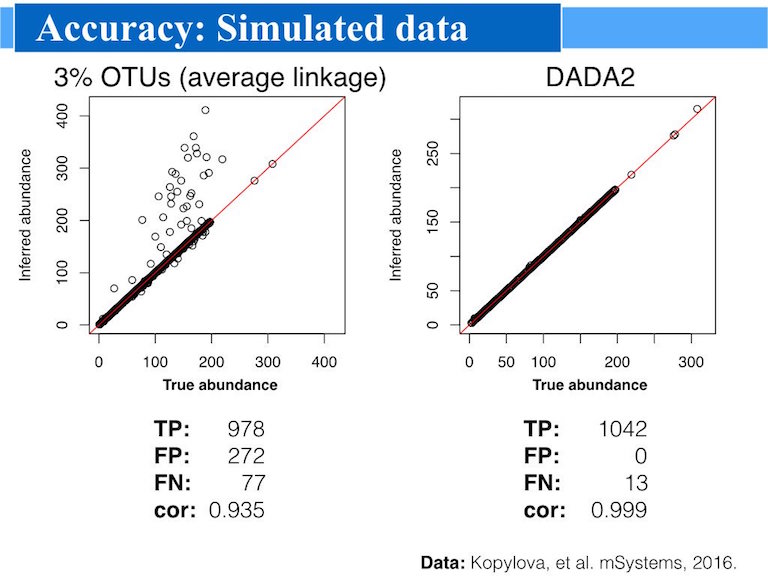
DADA2 implements a novel algorithm that models the errors introduced during amplicon sequencing, and uses that error model to infer the true sample composition. DADA2 replaces the traditional “OTU-picking” step in amplicon sequencing workflows, producing instead higher-resolution tables of amplicon sequence variants (ASVs).
The DADA2 method is more sensitive and specific than traditional OTU methods: DADA2 detects real biological variation missed by OTU methods while outputting fewer spurious sequences. How replacing OTUs with ASVs improves the precision, comprehensiveness and reproducibility of marker-gene data analysys 1.
--i-demultiplexed-seqs selects the demultiplexed input .qza files.
--p-trunc-len cuts all the reads to 150bp (depends on the quality of your reads).
Outputs:
--o-representative-sequences
--o-table table-dada2.qza \
--o-denoising-stats stats-dada2.qza
Code:
qiime dada2 denoise-single \
--i-demultiplexed-seqs demux-paired-end.qza \
--p-trim-left 0 \
--p-trunc-len 150 \
--o-representative-sequences rep-seqs-dada2.qza \
--o-table table-dada2.qza \
--o-denoising-stats stats-dada2.qza
Here is another way to perform dada2 analysis with R.
https://bioconductor.org/packages/devel/bioc/vignettes/dada2/inst/doc/dada2-intro.html