Software Design Document - CankayaUniversity/ceng-407-408-2019-2020-Image-Analysis-for-the-Classification-of-Brain-Tumor-Location-on-MR-Images GitHub Wiki
1. Introduction
I. Purpose
This software design document describes the architecture and system design of a project that planned to develop a method which detects brain tumor location on MR Images. During the past few years, brain tumor segmentation in Magnetic Resonance Imaging (MRI) has become an emergent research area in the field of medical imaging systems. Accurate detection of brain tumor plays a vital role in the diagnosis of brain cancer. In past decades many experiments has been done in medical ımage processing and artificial intelligence areas for detecting brain tumor, but no one algorithm or method outpeformed other methods. There is no algorithm found that works and detects the location of brain tumor %100 accurately and has a fast computation time. So, there is no one superior method. Accuracy and computation time are two important compenents for medical algorithms. Since, brain tumor is critical and sensitive topic, detection of tumor needs to be done properly and correctly. Over these experiments with algorithms, it is found that neural network methods and hybrid methods gives the best results considering computation and accuracy. Researchers , students ,professors and doctors who are interested in this field, wants to have more knowledge , wants to use or tests the method proposed can get benefits from this document.
II. Scope
For this project we want to work with medical images (MR Images) using image processing algorithms. We plan to use segmentation algorithms to detect whether brain tumor exists or not on brain MR Images. We plan to implement some of the methods used in past experiments in this field. We choose couple of experiments that gives the best results. We plan to implement these methods used on these experiments and compare their results depending on accuracy and computation time. We plan to combine these methods if possible and then we will try to improve the methods developed. This is the first phase of the project. If first base of the project becomes successful we will pass on to second phase and try to detect the type of the tumor. Second phase of the project is optional. We will also use preprocessing algorithms. MR Images carries a lot of noise which prevents the segmentation methods to work properly. These segmentation methods are used for detecting brain tumor region and noise decrease the efficiency of these methods working properly. So using preprocessing methods are necessary for this project. By using this method, doctors can get of their diagnosis verification and pre knowledge about the images. This project will also give contribution to medical image processing and artificial intelligence areas.
III. Glossary
Magnetic Resonance Imaging: Magnetic resonance imaging is a medical imaging technique used in radiology to form pictures of the anatomy and the physiological processes of the body.
Brain Tumor Segmentation: Brain tumor segmentation consists of separating the different tumor tissues (active tumor, edema and necrosis) from normal brain tissues.
Image processing: Image processing is a method to perform some operations on an image, in order to get an enhanced image or to extract some useful information from it. It is a type of signal processing in which input is an image and output may be image or characteristics/features associated with that image.
Image Segmentation: Image segmentation is the process of partitioning an image into multiple segments. Image segmentation is typically used to locate objects and boundaries in images.
Neural Networks: Neural networks are a set of algorithms, modeled loosely after the human brain, that are designed to recognize patterns.
Hybrid Method: Using more than one image processing algorithms on an image.
Artificial Intelligence:In computer science, artificial intelligence, sometimes called machine intelligence, is intelligence demonstrated by machines, in contrast to the natural intelligence displayed by humans.
IV. References
[1] IEEE Recommended Practice for Software Design Descriptions,2012, https://sovannarith.files.wordpress.com/2012/07/sdd_template.pdf
[2] IEEE Standard for Information Technology—Systems Design— Software Design Descriptions,2009 http://cengproject.cankaya.edu.tr/wp-content/uploads/sites/10/2017/12/SDD-ieee-1016-2009.pdf
V. Overview Of The Document
This document gives techinical details of the Project planned to detect tumor region on MR Images. The first part of this document gives an overall description about the project.Tın second part we gave detailed explanation of development of the project , design of the Project and planned Schedule for this project. In thirs part we gave techinal details of the method we will develop.
VI. Motivation
We are bunch of senior students in computer engineer department who are interested in image processing. Image processing has many fields. We want to improve ourselves on medical image processing. There is no, one algorithm that outperforms other algorithms on segmenting the medical images. Medical Image processing is a challenging and educational area, so we want to develop ourselves in this field. We also want to give good contribution to the world. Doctors can use this algorithms to be helpful for them on detecting brain tumors and start early treatment. This algorithms will be helpfull to doctors. We did very large research in this field before starting the Project, we also studied phyton programming language . phyton is the most suitable language for image processing since it has special libraries for image processing. Therefor, we will use phyton programming for this project.
2. DESIGN OVERVIEW
2.1 Description of Problem
There has been many image processing algorithms developed and many experiments done with these algorithms.Efficiency and accuracy of these algorithms are improved with these algorithms over the years. But, we still have long way to go to find the most successfull algorithms. Detection of required area is sensitive and critical subject in segmenting medical images. Accuracy and fast computation time is two important scales for these segmentation algorithms. Hybrid methods and neural network methods gave the best results during these experiments but still there is no algorithm or method that leaves out other methods. We plan to implement some of these algorithms and combine them, if it’s possible we also want to improve them. These methods will also help doctors in diagnosing brain tumors. Doctors will use these methods before diagnosing brain mr images and will have pre knowledge about the image and also they will get an second judgment about the image. This way their diagnose will be verified by a second person and the patients will start early treatment.
2.2 Technologies Used
We will use pyhton programming language, we will use Opencv libraries. We plan to use Visual Studio Code and Jupyter editors.We will use MySQL Server for storing database.There is no other technical tool needed for this project.
2.3 Architecture Design
2.3.1 Sequence Diagram
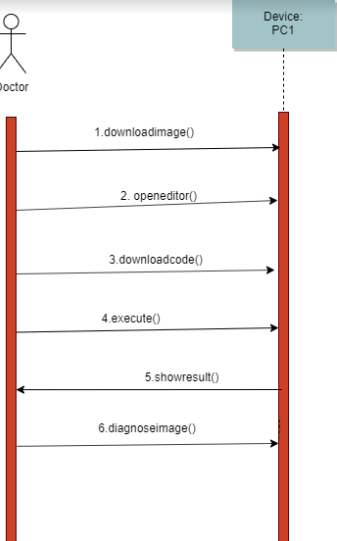
The Picture above shows the squence diagram of proposed method. The user needs have an already existed brain mr image in their computer. The user then needs to download the code of our code and needs to open it in an editor that Works with pyhton programming. The person than will execute the code and the system will Show the results of the image. The system will Show the original image and image that is segmented(that shows the area of tumor).
2.3.2 Activity Diagram
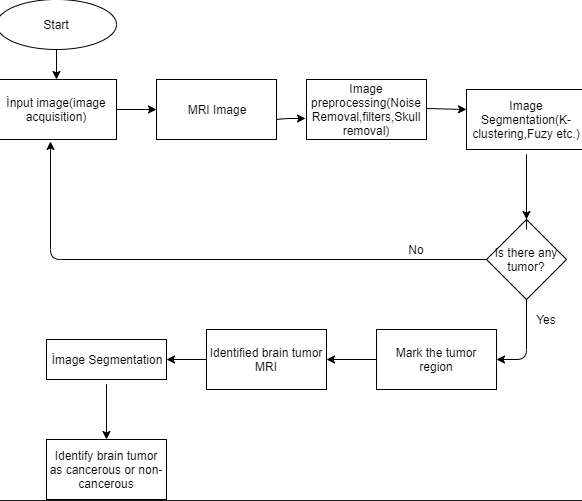
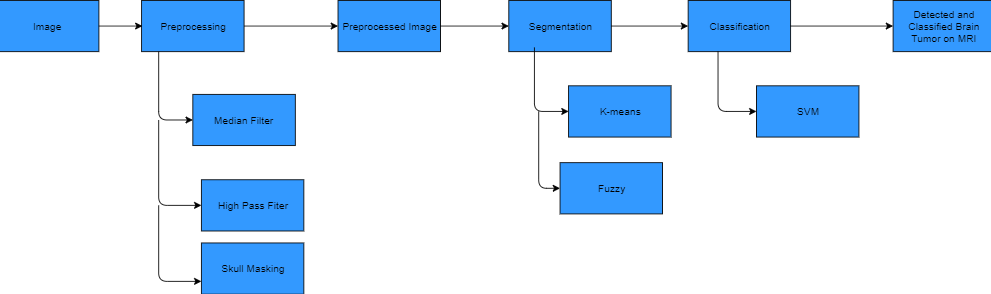
The figure above shows the activity diagram of system proposed. The user needs to put an image into the system which is called image acquisition. The image is then gonna be preprocessed to reduce the noise. The preprocessed algorithms are planned to be noise removal, filters and skull masking filter. After preprocessing, the system will use segmentation methods to detect tumor region. After using the segmentation methods, the system marks the tumor area. If there is no tumor found in the image, the system requires another image as an input. If these stages of the Project becomes succesfull; the system will implement another segmentation method that detects tumor type that detects tumor as cancerous or not cancerous.
2.3.3 Class Diagram
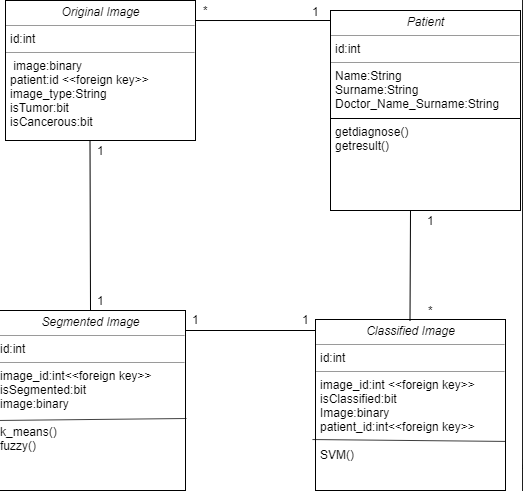
Class diagrams are used to show the classes, properties and relationships between each other and there are 4 classes in our project. We decided to store original images,segmented images and classified images at different classes so that it can be easier to implement the algorithms and show them to the user. We decided to not store preprocessed images, since preprocessing is only used to reduce nuise so that it can be easier to detect tumor region. Preprocessing is not directly related to detection of tumor. Therefor, we have 4 classes; Patient, Original Image, Segmentation Image, Classified Image. By Original Image, we mean original brain MR Images of the patients that has not been processed with any algorithms. The classes has id's as their priority keys, these priority keys defines different members of each class.The original image is directed to preprocessing than preprocessed image is directed to segmentation, then segmented image is directed to classification. This means that each original image has one segmented, classified and preprocessed image. Therefor, There is one-to-one relationship between the Original Images,Segmented Images and Classified Images. Classified ımages are results of the original ımages. Patient classes has one-to-many relationship with Original images and classified ımages since a patient can have more than one mr ımage.
2.3.4 Project Plan
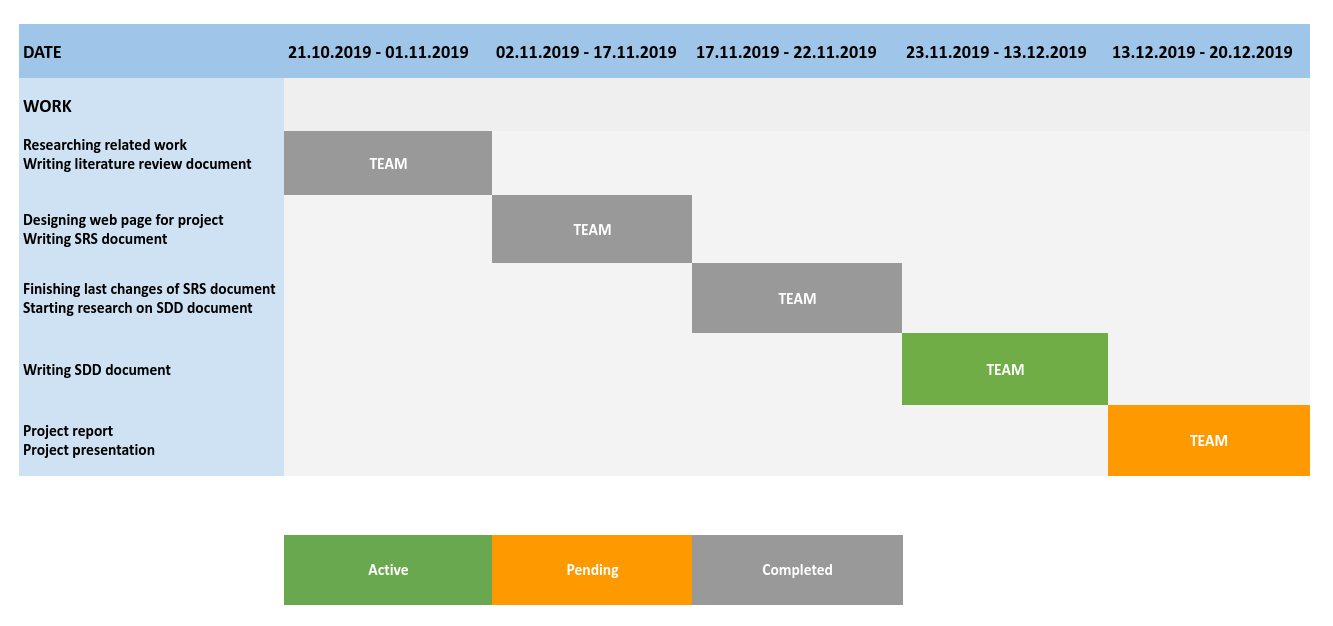
| Work | Description | Assignee(s) |
|---|---|---|
| Writing literature review document | Researching related literature, reading and documenting articles about the algorithms that will be used in the project | Team |
| Designing web page for project | Designing web page for publishing documents about project and team information | Team |
| Writing SRS document | Determining and documenting software requirements for project | Team |
| Writing SDD document | Identifying how data design and architecture design of software will be, documenting design description | Team |
| Project report | Collecting all documents about project in a single document | Team |
| Project presentation | Preparing presentation slide and making a demo presentation about what has been done so far | Team |
3.DATA DESIGN
3.1 Data Description
3.1.1 Entity Relationship (ER) Model
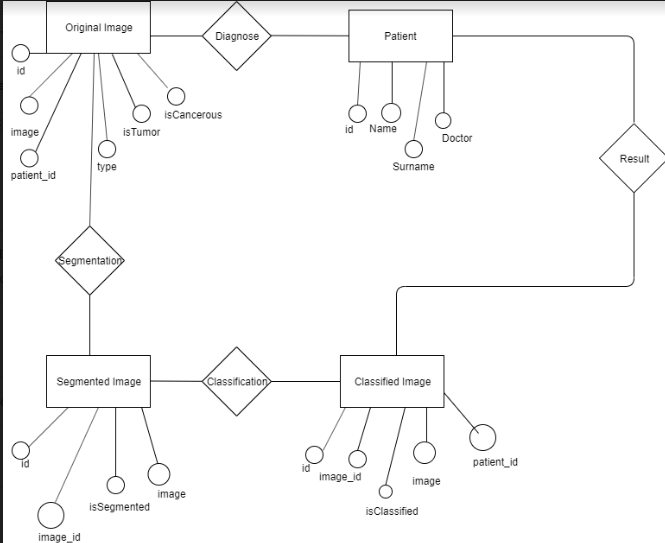
In the Figure, we showed the Image Analysis for the Classification of Brain Tumor Location on MR Images Entity Relationship Diagram. The ER Diagram shows Tables, Attributes, Relationships between tables will be used for database. The patients, original ımages, segmented images and classified images has id's as priority keys that defines them. Image_ıd attribute is a foreign key attribute for segmented and classified images that stores original image id. We store images as binary in these tables, so that it can be easier to show the images to users. We store type of the images on original image table. The brain mr images type is usually dicom but we will get the images from different sites so we want to also store them on type attribute on original image table. Original image has isTumor attribute as bit. It becomes 0 if there is no tumor detected on segmented image, it becomes 1 if there is tumor detected on segmented image. Original Image has, isCancerous attribute, it becomes 0 if there is no tumor detected or the tumor detected is found non-cancerous on classified image, it becomes 1 if detected tumor is found cancerous.We have isSegmented and isClassified atrributes on Segmented ımage and classified image tables. isSegmented attribute becomes 1 after segmentation is complete, isClassified image becomes 1 after classification is complete. This way the sytem will be ensured to follow the steps accordingly and not to implement classification before segmentation.
We will use MySql database server for storing database, since it is easy to use with python programming
3.2 Data
The data to be used on the system should be brain MR images. Mr image types are usually dicom and we expect to use dicom mr images. Dıcom images usually stores the tissue of the image, type of the image and information about the patient in it's format. DICOM (Digital Imaging and Communications in Medicine) is a standard for handling, storing, printing, and transmitting information in medical imaging. It includes a file format definition and a network communications protocol.[1]. We plan to get the MR images to use on the system and to implement algorithms from TCIA's[2], BRATS's[3] and kaggle's [4] websites.
4.USE CASE REALIZATION
4.1 Project Components
4.1.1 Brief Description of Project Components
From the research we did before starting this project, we picked 3 articles as references to implement for our project. These articles explain the details very well and very understandable and explains easy to implelement algorithms. These articles mostly use k-means, fuzzy and svm. From the research we did, it shows that hybrid method with SVM shows almost as good result as Neural Networks. Therefore, we decided to use K-means and Fuzzy(Hybrid Method) and SVM. The articles we choose as references are: [12][13][14]
4.1.1.1 Ready Packages
Ready packages in Python which will be used for machine learning algorithms (clustering, classification) and for image processing are listed as below:
- Numpy: for multi-dimensional array and matrix processing (image).
- Scipy: for optimization, linear algebra, integration and statistics.
- Scikit-learn: for machine learning algorithms, supports most of the supervised and unsupervised learning algorithms (clustering and classification).
- Matplotlib: for data visualization.
- Pydicom: for dealing with MRI images.
4.1.1.2 Image Preprocessing
Pre-processing is a common name for operations with images at the lowest level of abstraction; both input and output are intensity images.The aim of pre-processing is an improvement of the image data that suppresses unwanted distortions or enhances some image features important for further processing(segmentation). [5] In this project, preprocessing algorithms are used to reduce noise and distortions so that, segmentation algorithms can detect tumor region more accurately. We plan to use 3 preprocessing algorithms for this project; Median Filter, High Pass Filter, Skull Masking.
Median Filter
The Median Filter is a non-linear digital filtering technique, often used to remove noise from an image or signal. Such noise reduction is a typical pre-processing step to improve the results of later processing.It is particularly effective at removing ‘salt and pepper’ type noise.The median filter works by moving through the image pixel by pixel,replacing each value with the median value of neighbouring pixels.The pattern of neighbours is called the "window", which slides, pixel by pixel over the entire image. The median is calculated by first sorting all the pixel values from the window into numerical order, and then replacing the pixel being considered with the middle (median) pixel value. [6][7]
High Pass Filter
High pass filters are also called sharpening. A high-pass filter is a filter that passes high frequencies well, but attenuates frequencies lower than the cut-off frequency. Sharpening is fundamentally a highpass operation in the frequency domain. There are several standard forms of highpass filters such as Ideal, Butterworth and Gaussian highpass.Gaussian filter resulted better at sharpening the images filter in past experiments. A high-pass filter can be used to make an image appear sharper. [8]
Skull Masking
The quantitative morphometric studies of MR brain images often require a preliminary processing to isolate the brain from extra-cranial or non-brain tissues from MRI head scans, commonly referred to as skull stripping. Because the brain images that have preprocessed with automatic skull stripping eventually lead to get better segmentation of different brain regions which results for accurate diagnosis of various brain-related diseases. The brain regions must be skull-stripped prior to the application of other image processing algorithms. Moreover, skull stripping being a preliminary step, designed to eliminate non-brain tissues from MR brain images for many clinical applications and analyses, its accuracy and speed are considered as the key factors in the brain image segmentation and analysis. However, the accurate and automated skull stripping methods help to improve the speed and accuracy of prognostic and diagnostic procedures in medical applications. [9]
4.1.1.3 Image Segmentation
In computer vision, image segmentation is the process of partitioning a digital image into multiple segments. The goal of segmentation is to simplify and/or change the representation of an image into something that is more meaningful and easier to analyze.[15]
K-Means
K-means clustering algorithm is the simplest unsupervised learning algorithm that can solve clustering problem. The procedure followed to classify a given set of data through a certain number of clusters is very simple. In K-means ‘K’ centres are defined, one for each cluster. These clusters must be placed far away from each other. The next step is to take a point belonging to a given data set and associate it to the nearest centre. When no point is pending, the first step is completed and early grouping is done. The second step is to recalculate ‘k’ new centroids as barycentre of the clusters resulting from the previous step. After having ‘K’ new centroids a new binding has to be done between the same data set points and the nearest new centre. A loop has been generated. As a result of this loop, the k centres change their location step by step until centres do not move any more.[10]
Fuzzy C-Means
FCM clustering is an unsupervised method for the data analysis. This algorithm assigns membership to each data point corresponding to each cluster centre on the basis of distance between the cluster centre and the data point. The data point near to the cluster centre has more membership towards the particular centre. Generally, the summation of membership of each data point should be equal to one [10].
4.1.1.4 Image Classification
Image classification refers to a process in computer vision that can classify an image according to its visual content. For example, an image classification algorithm may be designed to tell if an image contains a human figure or not.[16]
SVM
Instead of minimizing an objective function based on the training samples (such as mean square error), the SVM attempts to minimize the bound on the generalization error (i.e., the error made by the learning machine on the test data not used during training). As a result, an SVM tends to perform well when applied to data outside the training set. SVM achieves this advantage by focusing on the training examples that are most difficult to classify. These “borderline” training examples are called support vectors. The SVM classifier is based on the hyperplane that maximizes the separating margin between the two classes. The basic SVM takes a set of input data and predicts, for each given input, which of two possible classes forms the output, making it a non-probabilistic binary linear classifier. In addition to performing linear classification, SVMs can efficiently perform a non-linear classification using kernel trick, implicitly mapping their inputs into high-dimensional feature spaces. To maximize the margin between the classes its kernel is used to control the empirical risk and classification. There are many kernel functions such as linear, polynomial of degree and Radial basis function (RBF). Among these kernel functions, a radial basis function proves to be better for MRI brain images [11].
SVM has two stages; training and testing stage. SVM trains itself by features given as an input to its learning algorithm. During training SVM selects the suitable margins between two classes. Features are labeled according to class associative with particular class. Artificial neural network has a few issues having local minima and number of neurons selection for each problem. In order to resolve this problem SVM occupies no local minima and overhead to neurons selection by initiating the idea of hyper planes [10].
5.REFERENCES
[1] DICOM Library https://www.dicomlibrary.com/dicom/
[2] TCIA(The Cancer Imaging Archieve) https://www.cancerimagingarchive.net/
[3] BRATS(Brain Tumor Segmentation) http://braintumorsegmentation.org/
[4] kaggle https://www.kaggle.com/
[5] https://www.slideshare.net/ASHI14march/image-pre-processing
[6] https://en.wikipedia.org › wiki › Median_filter
[7] https://www.cs.auckland.ac.nz/courses/compsci373s1c/PatricesLectures/Image%20Filtering_2up.pdf
[8] https://pdfs.semanticscholar.org/9b10/febd207e92f9d362ed06286d0ba31e290fec.pdf
[9] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4879034/
[10] MRI BRAIN IMAGES SEGMENTATION TECHNIQUES – A REVİEW, D.SELVARAJ Department of ECE, Sathyabama University, Chennai, Tamilnadu, India R.DHANASEKARAN, Syed Ammal Engineering College, Ramanathapuram, Tamilnadu, India, D.Selvaraj et.al / Indian Journal of Computer Science and Engineering (IJCSE), Vol. 4 No.5 Oct-Nov 2013
[11] Zhang, K., CAO, H.X., Yan, H (2006): “Application of support vector machines on network abnormal intrusion detection”, Application Research of Computers, Vol.5, pp.98-100.
[12] Mukambika P. S., Uma Rani K. IRJET. Segmentation and Classification of MRI Brain Tumor
[13] Mamidi Srujana , Mudadla Chandini , Ijjirythu Lavanya , R. Divyakanthi. Tumor Identification in Brain MRI Images Using Fuzzy-C-Means Algorithm
[14] S. R. Telrandhe, A. Pimpalkar, A. Kendhe (2016). Detection of Brain Tumor from MRI images by using Segmentation & SVM.
[15]https://en.wikipedia.org/wiki/Image_segmentation
[16]https://www.sciencedirect.com/topics/computer-science/image-classification