models Prov GigaPath - Azure/azureml-assets GitHub Wiki
Digital pathology poses unique computational challenges, as a standard gigapixel slide may comprise tens of thousands of image tiles^1,^2,^3. Previous models often rely predominantly on tile-level predictions, which can overlook critical slide-level context and spatial dependencies^4. Here we present Prov-GigaPath, a whole-slide pathology foundation model pretrained on 1.3 billion 256 × 256 pathology image tiles in 171,189 whole slides from Providence, a large U.S. health network comprising 28 cancer centers.
To pretrain Prov-GigaPath, we propose GigaPath, a novel vision transformer architecture for pretraining gigapixel pathology slides. To scale GigaPath for slide-level learning with tens of thousands of image tiles, GigaPath adapts the newly developed LongNet[^5] method to digital pathology.
For additional details, please see the publication: A whole-slide foundation model for digital pathology from real-world data
For documentation and example Jupyter Notebooks, visit: Prov-GigaPath - GitHub
Prov-GigaPath processes an entire histopathology slide by analyzing individual tiles and generating semantically meaningful embedding. These embeddings can be used as features for a wide range of clinical applications. Prov-GigaPath excels in long-context modelling of gigapixel pathology slides, by distilling varied local pathological structures and integrating global signatures across the whole slide. Prov-GigaPath consists of a tile encoder for capturing local features and a slide encoder for capturing global features. The tile encoder individually projects all tiles into compact embeddings. The slide encoder then inputs the sequence of tile embeddings and generates contextualized embeddings taking into account the entire sequence using a transformer. The tile encoder is pretrained using DINOv2, the state-of-the-art image self-supervised learning framework. The slide encoder combines masked autoencoder pretraining with LongNet5, our recently developed method for ultra long-sequence modelling. In downstream tasks, the output of the slide encoder is aggregated using a simple softmax attention layer.
The License for Prov-GigaPath is a research-use-only license: prov-gigapath/LICENSE.
The model is not intended or made available for clinical use as a medical device, clinical support, diagnostic tool, or other technology intended to be used in the diagnosis, cure, mitigation, treatment, or prevention of disease or other conditions. The model is not designed or intended to be a substitute for professional medical advice, diagnosis, treatment, or judgment and should not be used as such. All users are responsible for reviewing the output of the developed model to determine whether the model meets the user’s needs and for validating and evaluating the model before any clinical use.
For questions or comments, please contact: [email protected]
| Training Dataset | Details |
|---|---|
| Proprietary Datasets | Pretrained on 1.3 billion 256 × 256 pathology image tiles (20x, 0.5 MPP) in 171,189 whole slides from Providence. The slides originated from more than 30,000 patients covering 31 major tissue types. |
To evaluate Prov-GigaPath, we construct a digital pathology benchmark comprising 9 cancer subtyping tasks and 17 pathomics tasks, using both Providence and TCGA data. With large-scale pretraining and ultra-large-context modelling, Prov-GigaPath attains state-of-the-art performance on 25 out of 26 tasks, with significant improvement over the second-best method on 18 tasks.
| Task | Prov-GigaPath | HIPT | CtransPath | REMEDIS | p-value |
|---|---|---|---|---|---|
| NSCLC Typing | 0.756 ± 0.010 | 0.657 ± 0.013 | 0.732 ± 0.014 | 0.570 ± 0.015 | 0.065 |
| BRCA Typing | 0.899 ± 0.015 | 0.823 ± 0.027 | 0.895 ± 0.017 | 0.838 ± 0.025 | 0.539 |
| RCC Typing | 0.953 ± 0.007 *** | 0.900 ± 0.012 | 0.919 ± 0.011 | 0.804 ± 0.025 | 0.001 |
| COADREAD Typing | 0.834 ± 0.009* | 0.774 ± 0.013 | 0.799 ± 0.011 | 0.724 ± 0.018 | 0.014 |
| HB Typing | 0.905 ± 0.015* | 0.857 ± 0.023 | 0.858 ± 0.016 | 0.849 ± 0.013 | 0.01 |
| DIFG Typing | 0.970 ± 0.003 ** | 0.943 ± 0.008 | 0.945 ± 0.007 | 0.885 ± 0.011 | 0.005 |
| OVT Typing | 0.978 ± 0.003 *** | 0.946 ± 0.006 | 0.942 ± 0.007 | 0.834 ± 0.022 | 0.001 |
| CNS Typing | 0.956 ± 0.003 *** | 0.902 ± 0.006 | 0.922 ± 0.005 | 0.808 ± 0.019 | 0.01 |
| EGC Typing | 0.874 ± 0.011 | 0.857 ± 0.011 | 0.868 ± 0.013 | 0.832 ± 0.013 | 0.423 |
| Pan EGFR | 0.675 ± 0.011 *** | 0.637 ± 0.009 | 0.537 ± 0.006 | 0.613 ± 0.008 | 0.001 |
| Pan FAT1 | 0.648 ± 0.008 | 0.650 ± 0.005 | 0.646 ± 0.007 | 0.638 ± 0.006 | 0.652 |
| Pan KRAS | 0.775 ± 0.004 *** | 0.700 ± 0.006 | 0.642 ± 0.010 | 0.691 ± 0.009 | 0.001 |
| Pan LRP1B | 0.678 ± 0.006 *** | 0.644 ± 0.008 | 0.639 ± 0.009 | 0.638 ± 0.008 | 0.001 |
| Pan TP53 | 0.724 ± 0.006 *** | 0.653 ± 0.008 | 0.615 ± 0.006 | 0.679 ± 0.009 | 0.001 |
| LUAD EGFR | 0.543 ± 0.011* | 0.494 ± 0.012 | 0.510 ± 0.012 | 0.511 ± 0.011 | 0.032 |
| LUAD FAT1 | 0.712 ± 0.012* | 0.682 ± 0.014 | 0.688 ± 0.009 | 0.671 ± 0.019 | 0.024 |
| LUAD KRAS | 0.547 ± 0.008* | 0.536 ± 0.008 | 0.508 ± 0.014 | 0.532 ± 0.010 | 0.042 |
| LUAD LRP1B | 0.688 ± 0.014 | 0.655 ± 0.012 | 0.683 ± 0.013 | 0.651 ± 0.014 | 0.348 |
| LUAD TP53 | 0.638 ± 0.015* | 0.612 ± 0.012 | 0.614 ± 0.012 | 0.607 ± 0.016 | 0.042 |
| LUAD EGFR (TCGA) | 0.766 ± 0.012 ** | 0.606 ± 0.015 | 0.541 ± 0.016 | 0.619 ± 0.014 | 0.002 |
| LUAD FAT1 (TCGA) | 0.552 ± 0.021 | 0.466 ± 0.012 | 0.503 ± 0.015 | 0.523 ± 0.032 | 0.216 |
| LUAD KRAS (TCGA) | 0.610 ± 0.012 | 0.596 ± 0.010 | 0.472 ± 0.014 | 0.578 ± 0.006 | 0.188 |
| LUAD LRP1B (TCGA) | 0.598 ± 0.014 ** | 0.553 ± 0.010 | 0.529 ± 0.012 | 0.553 ± 0.014 | 0.01 |
| LUAD TP53 (TCGA) | 0.749 ± 0.011 *** | 0.679 ± 0.014 | 0.650 ± 0.016 | 0.702 ± 0.011 | 0.001 |
| Pan 18-biomarkers | 0.649 ± 0.003 *** | 0.626 ± 0.003 | 0.600 ± 0.002 | 0.628 ± 0.003 | 0.001 |
| Pan TMB | 0.708 ± 0.008 | 0.657 ± 0.010 | 0.695 ± 0.008 | 0.676 ± 0.008 | 0.097 |
Table comparing Prov-GigaPath with state-of-the-art pathology foundation models on 26 tasks in pathomics and cancer subtyping using AUROC. * indicates the significance level that Prov-GigaPath outperforms the best comparison approach on the specific task, with Wilcoxon test p-value< 5 x 10-2 for *, p-value<1x10-2 for ** , p-value< 1x 10-3 for ***. The last column shows the p-value using the one-sided Wilcoxon test.
The paper showcases a handful of supplementary figures which highlight the demographic statistics of the training population.
| Sex | % Patients |
|---|---|
| Female | 50.42% |
| Male | 49.50% |
| None | 0.08% |
Table: Sex distribution of patients in Prov-Path.
| Age | % Patients |
|---|---|
| Below 11 | 0.21% |
| 11-20 | 0.25% |
| 21-30 | 1.09% |
| 31-40 | 2.99% |
| 41-50 | 8.70% |
| 51-60 | 23.48% |
| 61-70 | 32.37% |
| 71-80 | 21.89% |
| 81-90 | 13.43% |
| 91-100 | 0.80% |
Table: Age distribution of patients in Prov-Path.
| Race | % Patients |
|---|---|
| White or Caucasian | 78.28% |
| Asian | 4.31% |
| Black or African American | 1.83% |
| American Indian or Alaska Native | 0.76% |
| Native Hawaiian or Other Pacific Islander | 0.33% |
| Unknown | 8.20% |
| Patient Refused | 1.97% |
| Other | 4.32% |
Table: Self-reported ethnicity distribution of patients in Prov-Path.
Responsible AI is a shared responsibility and we have identified six principles and practices help organizations address risks, innovate, and create value: fairness, reliability and safety, privacy and security, inclusiveness, transparency, and accountability. When downloaded or used in accordance with our terms of service, developers should work with their supporting model team to ensure this model meets requirements for the relevant use case and addresses unforeseen product misuse. While testing the model with images and/or text, ensure the the data is PHI free and that there is no patient information or information that can be tracked to a patient identity.
The data, code, and model checkpoints are intended to be used solely for (I) future research on pathology foundation models and (II) reproducibility of the experimental results reported in the reference paper. The data, code, and model checkpoints are not intended to be used in clinical care or for any clinical decision-making purposes.
The primary intended use is to support AI researchers reproducing and building on top of this work. GigaPath should be helpful for exploring pre-training, and encoding of digital pathology slides data.
- Any deployed use case of the model — commercial or otherwise — is out of scope. Although we evaluated the models using a broad set of publicly-available research benchmarks, the models and evaluations are intended for research use only and not intended for deployed or clinical use cases.
- Use by clinicians to inform clinical decision-making, as a diagnostic tool, or as a medical device — GigaPath is not designed or intended to be deployed as-is in clinical settings nor is it for use in the diagnosis, cure, mitigation, treatment, or prevention of disease or other conditions (including to support clinical decision-making), or as a substitute of professional medical advice, diagnosis, treatment, or clinical judgment of a healthcare professional.
- Scenarios without consent for data — Any scenario that uses health data for a purpose for which consent was not obtained.
- Use outside of health scenarios — Any scenario that uses non-medical related image and/or serving purposes outside of the healthcare domain.
Please see Microsoft's Responsible AI Principles and approach available at Microsoft's Responsible AI Principles.
data = {
"input_data": {
"columns": ["image"],
"index": list(range(len(image_paths))),
"data": [
[
base64.encodebytes(read_image(path)).decode("utf-8")
] for path in image_paths
]
}
}Output: Outputs from the API are image embeddings users can do various downstream tasks, including PCA analysis (example below).
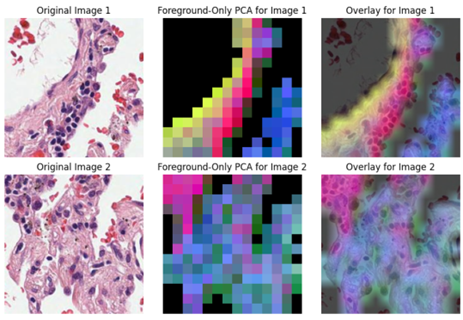
A de-identified sample subset of the Prov-Path data can be accessed from these links [^5],[^6].
Sample notebooks can be accessed below as well. They assume a HuggingFace distribution of the model:
- Semantic visualizations of the GigaPath tile embeddings
- Calculating GigaPath slide-level embeddings
- Fine-tuning GigaPath for downstream tasks
- Supports: CPU and GPU
- Default: Single V100 GPU or Intel CPU
- Minimum: Single GPU instance with 8GB memory (fastest) or CPU instance
[^5]https://zenodo.org/records/10909616 [^6]: https://zenodo.org/records/10909922
For more information on responsible AI practices, refer to Microsoft's Responsible AI Principles at https://www.microsoft.com/en-us/ai/principles-and-approach/.
Version: 3
task : image-feature-extraction industry : health-and-life-sciences Preview inference_supported_envs : ['hf'] license : mit author : Microsoft hiddenlayerscanned SharedComputeCapacityEnabled inference_compute_allow_list : ['Standard_NC6s_v3', 'Standard_NC12s_v3', 'Standard_NC24s_v3', 'Standard_NC24ads_A100_v4', 'Standard_NC48ads_A100_v4', 'Standard_NC96ads_A100_v4', 'Standard_ND96asr_v4', 'Standard_ND96amsr_A100_v4', 'Standard_ND40rs_v2']
View in Studio: https://ml.azure.com/registries/azureml/models/Prov-GigaPath/version/3
License: mit
inference-min-sku-spec: 6|1|112|64
inference-recommended-sku: Standard_NC6s_v3, Standard_NC12s_v3, Standard_NC24s_v3, Standard_NC24ads_A100_v4, Standard_NC48ads_A100_v4, Standard_NC96ads_A100_v4, Standard_ND96asr_v4, Standard_ND96amsr_A100_v4, Standard_ND40rs_v2
languages: en
SharedComputeCapacityEnabled: True